nnUNet(v1)框架使用全过程讲解
nnUNet作为医学图像分割的框架去使用十分的方便,所以可以简单地学习了解一下它的代码。这里只讲部分重要代码以及使用。
注:介绍的是nnunetv1.7版本,因为作者的条件限制,目前只能使用v1版本,虽然比较古老且代码条理不够清晰,但也够用,还请见谅。
关于nnunet的介绍以及简单使用可以看博主的这两个文章:
首先是下载代码到本地,不要自己乱找,因为作者第一次虽然成功跑通了代码,但是在后续跑其他数据集的时候发现代码的版本问题很混乱,所以推荐用这个下载:
cd /...
wget https://github.com/MIC-DKFZ/nnUNet/archive/refs/tags/v1.7.0.zip
unzip v1.7.0.zip
#解压后改一下名字nnuent或者其他都行
cd /.../nnunet/nnUNet
pip install -e .下面我先再次介绍一下两个重要的步骤:预处理数据与训练。
数据预处理
由我的nnunet基础使用的文章可以知道我的预处理数据命令是:
nnUNet_plan_and_preprocess -t 500 -tl 8 注:如果不pip install -e .的话这里是无法使用nnUNet_plan_and_preprocess类似的命令的,报错类似于:
root@d21s9hmvd8ehu-0:/xujiheng# export nnUNet_raw_data_base="/xujiheng/Synapse/nnUNet/nnUNet/nnUNetFrame/DATASET/nnUNet_raw"
root@d21s9hmvd8ehu-0:/xujiheng# export nnUNet_preprocessed="/xujiheng/Synapse/nnUNet/nnUNet/nnUNetFrame/DATASET/nnUNet_preprocessed"
root@d21s9hmvd8ehu-0:/xujiheng# export RESULTS_FOLDER="/xujiheng/Synapse/nnUNet/nnUNet/nnUNetFrame/DATASET/nnUNet_trained_models"
root@d21s9hmvd8ehu-0:/xujiheng#
root@d21s9hmvd8ehu-0:/xujiheng# nnUNet_plan_and_preprocess -t 2
Traceback (most recent call last):
File "/usr/local/bin/nnUNet_plan_and_preprocess", line 33, in <module>
sys.exit(load_entry_point('nnunet', 'console_scripts', 'nnUNet_plan_and_preprocess')())
File "/usr/local/bin/nnUNet_plan_and_preprocess", line 22, in importlib_load_entry_point
for entry_point in distribution(dist_name).entry_points
File "/usr/lib/python3.8/importlib/metadata.py", line 445, in distribution
return Distribution.from_name(distribution_name)
File "/usr/lib/python3.8/importlib/metadata.py", line 169, in from_name
raise PackageNotFoundError(name)
importlib.metadata.PackageNotFoundError: nnunet但是可以使用python命令:
cd /xujiheng/nnunet/nnUNet
python -m nnunet.experiment_planning.nnUNet_plan_and_preprocess -t 2那它到底做了什么?
这是 nnU-Net 自动化 pipeline 的核心第一步,分为两个阶段:planning(规划) + preprocessing(预处理)。
-t 500:指定任务 ID
对应数据集目录:Task500_Synapse。
nnU-Net 会去 nnUNet_raw_data_base/nnUNet_raw_data/Task500_... 读取 dataset.json 和图像/标签。
Planning(规划)阶段做了什么?
nnU-Net 会分析你的 8 个训练样本,自动决定:
|
决策项 |
说明 |
|---|---|
|
目标 spacing(体素间距) |
根据图像各向异性程度,决定是否重采样到统一分辨率(如 [1.5, 1.5, 2.0] mm) |
|
输入 patch size |
根据 GPU 显存和图像大小,自动选择 3D patch 尺寸(如 [128, 128, 128]) |
|
是否启用 3D_lowres |
如果原始图像太大(如 > 200³),会额外生成低分辨率版本用于 cascade 模型 |
|
强度归一化方式 |
CT 用固定窗宽窗位(如 -1000~1000),MRI 用 per-case z-score |
输出文件:
$nnUNet_preprocessed/Task500_.../nnUNetPlansv2.1_plans_3D.pkl
Preprocessing(预处理)阶段做了什么?
对每个训练样本执行:
(1)重采样:将图像和标签插值到 planning 阶段确定的统一 spacing
(2)强度归一化:clip 到 [-1000, 1000],再归一化到 [0, 1]
(3)前景区域裁剪:只保留包含器官的 bounding box(大幅减少无效背景)
(4)保存为 .npy 格式:加速训练时的数据加载
总结一下:
运行 nnUNet_plan_and_preprocess -t 500 -tl 8 后,nnU-Net 会基于你提供的 8 个训练样本,自动完成数据规划(planning)与预处理(preprocessing),最终生成以下关键内容:
(1)标准化的预处理数据:将原始 .nii.gz 图像和标签重采样到统一各向同性 spacing、进行强度归一化(如 CT 窗宽窗位裁剪)、前景区域裁剪,并保存为高效的 .npy 格式;
(2)训练配置文件(plans.pkl):Python 字典(dict),用 pickle 序列化保存,包含针对该数据集自动推导出的网络输入尺寸(patch_size)、体素间距(spacing)、batch size、U-Net 下采样结构等超参数;
(3)可用模型类型决策:根据图像大小和 GPU 显存估算,决定是否启用 2d、3d_fullres 或 3d_lowres 等训练方案。
训练
我的训练的代码是:
export nnUNet_raw_data_base="/xujiheng/Synapse/nnUNet/nnUNet/nnUNetFrame/DATASET/nnUNet_raw" && export nnUNet_preprocessed="/xujiheng/Synapse/nnUNet/nnUNet/nnUNetFrame/DATASET/nnUNet_preprocessed" && export RESULTS_FOLDER="/xujiheng/Synapse/nnUNet/nnUNet/nnUNetFrame/DATASET/nnUNet_trained_models" && python /xujiheng/Synapse/nnUNet/nnUNet/nnunet/run/run_training.py 3d_fullres nnUNetTrainerV2 500 all| 环境变量 | 存什么 | 谁生成 | 谁使用 |
|---|---|---|---|
nnUNet_raw_data_base |
原始 .nii.gz 数据 |
手动准备 | nnUNet_plan_and_preprocess |
nnUNet_preprocessed |
预处理后的 .npy + 配置 |
nnUNet_plan_and_preprocess |
run_training.py(训练) |
RESULTS_FOLDER |
模型权重、日志 | run_training.py |
(查看结果)、推理脚本 |
| 参数 | 含义 | 说明 |
|---|---|---|
3d_fullres |
网络配置类型 | 表示使用预处理阶段生成的“全分辨率 3D U-Net”方案(对应 plans_3D.pkl 中的 stage) |
nnUNetTrainerV2 |
训练器类 | 默认训练器,包含: Dice + CE loss 强数据增强(旋转、缩放、弹性形变等) 学习率调度(poly decay) 自动验证 |
500 |
任务 ID | 对应 Task500_YourName,nnU-Net 会自动拼接路径查找数据 |
all |
交叉验证 fold | all 表示使用全部 8 个样本训练,不划分验证集(常用于最终模型训练)若写 0~4,则进行 5 折交叉验证 |
现在来关注一下上面的数据预处理与训练的关系:
预处理的输出直接决定了后续训练的可行性与配置:例如使用的 3d_fullres 模式,其网络结构、输入尺寸、数据增强策略等全部源自预处理阶段生成的 nnUNetPlansv2.1_plans_3D.pkl 文件。
没有这一步,训练无法启动;而一旦完成,训练命令只需指定 3d_fullres,框架便会自动加载对应的预处理数据和模型配置,实现“数据驱动的全自动训练”。
nnU-Net 会根据8 个训练样本的图像尺寸、spacing、GPU 显存估算,自动决定是否生成以下三种模型配置("plans"):
2d:2D U-Net(逐 slice 训练)
3d_lowres:低分辨率 3D U-Net(用于大图像)
3d_fullres:全分辨率 3D U-Net(标准配置)
代码部分
nnUNet_plan_and_preprocess(规划与预处理)
代码位置如下:
# Copyright 2020 Division of Medical Image Computing, German Cancer Research Center (DKFZ), Heidelberg, Germany
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import nnunet
from batchgenerators.utilities.file_and_folder_operations import *
from nnunet.experiment_planning.DatasetAnalyzer import DatasetAnalyzer
from nnunet.experiment_planning.utils import crop
from nnunet.paths import *
import shutil
from nnunet.utilities.task_name_id_conversion import convert_id_to_task_name
from nnunet.preprocessing.sanity_checks import verify_dataset_integrity
from nnunet.training.model_restore import recursive_find_python_class
def main():
import argparse
parser = argparse.ArgumentParser()
parser.add_argument("-t", "--task_ids", nargs="+", help="List of integers belonging to the task ids you wish to run"
" experiment planning and preprocessing for. Each of these "
"ids must, have a matching folder 'TaskXXX_' in the raw "
"data folder")
parser.add_argument("-pl3d", "--planner3d", type=str, default="ExperimentPlanner3D_v21",
help="Name of the ExperimentPlanner class for the full resolution 3D U-Net and U-Net cascade. "
"Default is ExperimentPlanner3D_v21. Can be 'None', in which case these U-Nets will not be "
"configured")
parser.add_argument("-pl2d", "--planner2d", type=str, default="ExperimentPlanner2D_v21",
help="Name of the ExperimentPlanner class for the 2D U-Net. Default is ExperimentPlanner2D_v21. "
"Can be 'None', in which case this U-Net will not be configured")
parser.add_argument("-no_pp", action="store_true",
help="Set this flag if you dont want to run the preprocessing. If this is set then this script "
"will only run the experiment planning and create the plans file")
parser.add_argument("-tl", type=int, required=False, default=8,
help="Number of processes used for preprocessing the low resolution data for the 3D low "
"resolution U-Net. This can be larger than -tf. Don't overdo it or you will run out of "
"RAM")
parser.add_argument("-tf", type=int, required=False, default=8,
help="Number of processes used for preprocessing the full resolution data of the 2D U-Net and "
"3D U-Net. Don't overdo it or you will run out of RAM")
parser.add_argument("--verify_dataset_integrity", required=False, default=False, action="store_true",
help="set this flag to check the dataset integrity. This is useful and should be done once for "
"each dataset!")
parser.add_argument("-overwrite_plans", type=str, default=None, required=False,
help="Use this to specify a plans file that should be used instead of whatever nnU-Net would "
"configure automatically. This will overwrite everything: intensity normalization, "
"network architecture, target spacing etc. Using this is useful for using pretrained "
"model weights as this will guarantee that the network architecture on the target "
"dataset is the same as on the source dataset and the weights can therefore be transferred.\n"
"Pro tip: If you want to pretrain on Hepaticvessel and apply the result to LiTS then use "
"the LiTS plans to run the preprocessing of the HepaticVessel task.\n"
"Make sure to only use plans files that were "
"generated with the same number of modalities as the target dataset (LiTS -> BCV or "
"LiTS -> Task008_HepaticVessel is OK. BraTS -> LiTS is not (BraTS has 4 input modalities, "
"LiTS has just one)). Also only do things that make sense. This functionality is beta with"
"no support given.\n"
"Note that this will first print the old plans (which are going to be overwritten) and "
"then the new ones (provided that -no_pp was NOT set).")
parser.add_argument("-overwrite_plans_identifier", type=str, default=None, required=False,
help="If you set overwrite_plans you need to provide a unique identifier so that nnUNet knows "
"where to look for the correct plans and data. Assume your identifier is called "
"IDENTIFIER, the correct training command would be:\n"
"'nnUNet_train CONFIG TRAINER TASKID FOLD -p nnUNetPlans_pretrained_IDENTIFIER "
"-pretrained_weights FILENAME'")
args = parser.parse_args()
task_ids = args.task_ids
dont_run_preprocessing = args.no_pp
tl = args.tl
tf = args.tf
planner_name3d = args.planner3d
planner_name2d = args.planner2d
if planner_name3d == "None":
planner_name3d = None
if planner_name2d == "None":
planner_name2d = None
if args.overwrite_plans is not None:
if planner_name2d is not None:
print("Overwriting plans only works for the 3d planner. I am setting '--planner2d' to None. This will "
"skip 2d planning and preprocessing.")
assert planner_name3d == 'ExperimentPlanner3D_v21_Pretrained', "When using --overwrite_plans you need to use " \
"'-pl3d ExperimentPlanner3D_v21_Pretrained'"
# we need raw data
tasks = []
for i in task_ids:
i = int(i)
task_name = convert_id_to_task_name(i)
if args.verify_dataset_integrity:
verify_dataset_integrity(join(nnUNet_raw_data, task_name))
crop(task_name, False, tf)
tasks.append(task_name)
search_in = join(nnunet.__path__[0], "experiment_planning")
if planner_name3d is not None:
planner_3d = recursive_find_python_class([search_in], planner_name3d, current_module="nnunet.experiment_planning")
if planner_3d is None:
raise RuntimeError("Could not find the Planner class %s. Make sure it is located somewhere in "
"nnunet.experiment_planning" % planner_name3d)
else:
planner_3d = None
if planner_name2d is not None:
planner_2d = recursive_find_python_class([search_in], planner_name2d, current_module="nnunet.experiment_planning")
if planner_2d is None:
raise RuntimeError("Could not find the Planner class %s. Make sure it is located somewhere in "
"nnunet.experiment_planning" % planner_name2d)
else:
planner_2d = None
for t in tasks:
print("\n\n\n", t)
cropped_out_dir = os.path.join(nnUNet_cropped_data, t)
preprocessing_output_dir_this_task = os.path.join(preprocessing_output_dir, t)
#splitted_4d_output_dir_task = os.path.join(nnUNet_raw_data, t)
#lists, modalities = create_lists_from_splitted_dataset(splitted_4d_output_dir_task)
# we need to figure out if we need the intensity propoerties. We collect them only if one of the modalities is CT
dataset_json = load_json(join(cropped_out_dir, 'dataset.json'))
modalities = list(dataset_json["modality"].values())
collect_intensityproperties = True if (("CT" in modalities) or ("ct" in modalities)) else False
dataset_analyzer = DatasetAnalyzer(cropped_out_dir, overwrite=False, num_processes=tf) # this class creates the fingerprint
_ = dataset_analyzer.analyze_dataset(collect_intensityproperties) # this will write output files that will be used by the ExperimentPlanner
maybe_mkdir_p(preprocessing_output_dir_this_task)
shutil.copy(join(cropped_out_dir, "dataset_properties.pkl"), preprocessing_output_dir_this_task)
shutil.copy(join(nnUNet_raw_data, t, "dataset.json"), preprocessing_output_dir_this_task)
threads = (tl, tf)
print("number of threads: ", threads, "\n")
if planner_3d is not None:
if args.overwrite_plans is not None:
assert args.overwrite_plans_identifier is not None, "You need to specify -overwrite_plans_identifier"
exp_planner = planner_3d(cropped_out_dir, preprocessing_output_dir_this_task, args.overwrite_plans,
args.overwrite_plans_identifier)
else:
exp_planner = planner_3d(cropped_out_dir, preprocessing_output_dir_this_task)
exp_planner.plan_experiment()
if not dont_run_preprocessing: # double negative, yooo
exp_planner.run_preprocessing(threads)
if planner_2d is not None:
exp_planner = planner_2d(cropped_out_dir, preprocessing_output_dir_this_task)
exp_planner.plan_experiment()
if not dont_run_preprocessing: # double negative, yooo
exp_planner.run_preprocessing(threads)
if __name__ == "__main__":
main()
nnU-Net 的核心优势就是“自动化”,而这一步正是实现自动化的起点。nnUNet_plan_and_preprocess 的作用是对一个或多个医学图像任务(TaskXXX),自动完成:
(1)数据完整性检查(可选)
(2)图像裁剪(crop)到前景区域
(3)分析数据集特征(如模态、尺寸、spacing、强度分布)
(4)生成“实验计划”(plans.pkl) —— 决定是否训练 2D / 3D_fullres / 3D_lowres
(5)执行预处理(重采样、归一化)并保存为 .npy 格式
只需提供原始 .nii.gz 数据和 dataset.json,nnU-Net 就能自动决定“怎么训”。它生成的 plans.pkl 是训练时的“蓝图”,包含网络结构、输入大小等关键参数。预处理后的 .npy 文件能大幅提升训练速度和稳定性。
run_training.py
nnU-Net 模型训练的主入口脚本,它的核心任务是:
(1)解析参数:输入网络类型(2D/3D/级联)、任务、fold、训练器名称等。
(2)加载配置:通过 get_default_configuration 读取由 nnUNet_plan_and_preprocess 生成的 plans.pkl,确定训练参数(如patch大小、batch size、使用的Trainer类等)。
(3)构建训练器(Trainer):实例化对应的 nnUNetTrainer 类(如 nnUNetTrainerV2),该类封装了训练全过程。
(4)初始化训练:设置数据加载器、网络结构、优化器、损失函数等。
(5)执行训练或验证:
若不是 --validation_only,则调用 trainer.run_training() 开始训练。否则加载训练好的模型权重,运行验证集推理。
(6)保存结果:保存模型检查点(checkpoint),并在训练结束后对验证集进行预测(可选保存 softmax 输出)。
(7)级联支持:如果是 3d_lowres 阶段,会自动调用 predict_next_stage 为 3d_cascade_fullres 阶段生成输入。
调用的关键模块:
| 模块 | 作用 |
|---|---|
get_default_configuration() |
读取 plans 文件,确定输出路径、数据路径、Trainer 类等 |
nnUNetTrainer 及其子类 |
核心训练逻辑:训练循环、损失函数、学习率策略、数据增强等 |
load_pretrained_weights() |
支持加载预训练权重(迁移学习) |
predict_next_stage() |
在 3D 级联流程中,用低分辨率模型预测高分辨率模型的输入 |
# Copyright 2020 Division of Medical Image Computing, German Cancer Research Center (DKFZ), Heidelberg, Germany
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import argparse
from batchgenerators.utilities.file_and_folder_operations import *
from nnunet.run.default_configuration import get_default_configuration
from nnunet.paths import default_plans_identifier
from nnunet.run.load_pretrained_weights import load_pretrained_weights
from nnunet.training.cascade_stuff.predict_next_stage import predict_next_stage
from nnunet.training.network_training.nnUNetTrainer import nnUNetTrainer
from nnunet.training.network_training.nnUNetTrainerCascadeFullRes import nnUNetTrainerCascadeFullRes
from nnunet.training.network_training.nnUNetTrainerV2_CascadeFullRes import nnUNetTrainerV2CascadeFullRes
from nnunet.utilities.task_name_id_conversion import convert_id_to_task_name
def main():
parser = argparse.ArgumentParser()
parser.add_argument("network")
parser.add_argument("network_trainer")
parser.add_argument("task", help="can be task name or task id")
parser.add_argument("fold", help='0, 1, ..., 5 or \'all\'')
parser.add_argument("-val", "--validation_only", help="use this if you want to only run the validation",
action="store_true")
parser.add_argument("-c", "--continue_training", help="use this if you want to continue a training",
action="store_true")
parser.add_argument("-p", help="plans identifier. Only change this if you created a custom experiment planner",
default=default_plans_identifier, required=False)
parser.add_argument("--use_compressed_data", default=False, action="store_true",
help="If you set use_compressed_data, the training cases will not be decompressed. Reading compressed data "
"is much more CPU and RAM intensive and should only be used if you know what you are "
"doing", required=False)
parser.add_argument("--deterministic",
help="Makes training deterministic, but reduces training speed substantially. I (Fabian) think "
"this is not necessary. Deterministic training will make you overfit to some random seed. "
"Don't use that.",
required=False, default=False, action="store_true")
parser.add_argument("--npz", required=False, default=False, action="store_true", help="if set then nnUNet will "
"export npz files of "
"predicted segmentations "
"in the validation as well. "
"This is needed to run the "
"ensembling step so unless "
"you are developing nnUNet "
"you should enable this")
parser.add_argument("--find_lr", required=False, default=False, action="store_true",
help="not used here, just for fun")
parser.add_argument("--valbest", required=False, default=False, action="store_true",
help="hands off. This is not intended to be used")
parser.add_argument("--fp32", required=False, default=False, action="store_true",
help="disable mixed precision training and run old school fp32")
parser.add_argument("--val_folder", required=False, default="validation_raw",
help="name of the validation folder. No need to use this for most people")
parser.add_argument("--disable_saving", required=False, action='store_true',
help="If set nnU-Net will not save any parameter files (except a temporary checkpoint that "
"will be removed at the end of the training). Useful for development when you are "
"only interested in the results and want to save some disk space")
parser.add_argument("--disable_postprocessing_on_folds", required=False, action='store_true',
help="Running postprocessing on each fold only makes sense when developing with nnU-Net and "
"closely observing the model performance on specific configurations. You do not need it "
"when applying nnU-Net because the postprocessing for this will be determined only once "
"all five folds have been trained and nnUNet_find_best_configuration is called. Usually "
"running postprocessing on each fold is computationally cheap, but some users have "
"reported issues with very large images. If your images are large (>600x600x600 voxels) "
"you should consider setting this flag.")
parser.add_argument("--disable_validation_inference", required=False, action="store_true",
help="If set nnU-Net will not run inference on the validation set. This is useful if you are "
"only interested in the test set results and want to save some disk space and time.")
# parser.add_argument("--interp_order", required=False, default=3, type=int,
# help="order of interpolation for segmentations. Testing purpose only. Hands off")
# parser.add_argument("--interp_order_z", required=False, default=0, type=int,
# help="order of interpolation along z if z is resampled separately. Testing purpose only. "
# "Hands off")
# parser.add_argument("--force_separate_z", required=False, default="None", type=str,
# help="force_separate_z resampling. Can be None, True or False. Testing purpose only. Hands off")
parser.add_argument('--val_disable_overwrite', action='store_false', default=True,
help='Validation does not overwrite existing segmentations')
parser.add_argument('--disable_next_stage_pred', action='store_true', default=False,
help='do not predict next stage')
parser.add_argument('-pretrained_weights', type=str, required=False, default=None,
help='path to nnU-Net checkpoint file to be used as pretrained model (use .model '
'file, for example model_final_checkpoint.model). Will only be used when actually training. '
'Optional. Beta. Use with caution.')
args = parser.parse_args()
task = args.task
fold = args.fold
network = args.network
network_trainer = args.network_trainer
validation_only = args.validation_only
plans_identifier = args.p
find_lr = args.find_lr
disable_postprocessing_on_folds = args.disable_postprocessing_on_folds
use_compressed_data = args.use_compressed_data
decompress_data = not use_compressed_data
deterministic = args.deterministic
valbest = args.valbest
fp32 = args.fp32
run_mixed_precision = not fp32
val_folder = args.val_folder
# interp_order = args.interp_order
# interp_order_z = args.interp_order_z
# force_separate_z = args.force_separate_z
if not task.startswith("Task"):
task_id = int(task)
task = convert_id_to_task_name(task_id)
if fold == 'all':
pass
else:
fold = int(fold)
# if force_separate_z == "None":
# force_separate_z = None
# elif force_separate_z == "False":
# force_separate_z = False
# elif force_separate_z == "True":
# force_separate_z = True
# else:
# raise ValueError("force_separate_z must be None, True or False. Given: %s" % force_separate_z)
plans_file, output_folder_name, dataset_directory, batch_dice, stage, \
trainer_class = get_default_configuration(network, task, network_trainer, plans_identifier)
if trainer_class is None:
raise RuntimeError("Could not find trainer class in nnunet.training.network_training")
if network == "3d_cascade_fullres":
assert issubclass(trainer_class, (nnUNetTrainerCascadeFullRes, nnUNetTrainerV2CascadeFullRes)), \
"If running 3d_cascade_fullres then your " \
"trainer class must be derived from " \
"nnUNetTrainerCascadeFullRes"
else:
assert issubclass(trainer_class,
nnUNetTrainer), "network_trainer was found but is not derived from nnUNetTrainer"
trainer = trainer_class(plans_file, fold, output_folder=output_folder_name, dataset_directory=dataset_directory,
batch_dice=batch_dice, stage=stage, unpack_data=decompress_data,
deterministic=deterministic,
fp16=run_mixed_precision)
if args.disable_saving:
trainer.save_final_checkpoint = False # whether or not to save the final checkpoint
trainer.save_best_checkpoint = False # whether or not to save the best checkpoint according to
# self.best_val_eval_criterion_MA
trainer.save_intermediate_checkpoints = True # whether or not to save checkpoint_latest. We need that in case
# the training chashes
trainer.save_latest_only = True # if false it will not store/overwrite _latest but separate files each
trainer.initialize(not validation_only)
if find_lr:
trainer.find_lr()
else:
if not validation_only:
if args.continue_training:
# -c was set, continue a previous training and ignore pretrained weights
trainer.load_latest_checkpoint()
elif (not args.continue_training) and (args.pretrained_weights is not None):
# we start a new training. If pretrained_weights are set, use them
load_pretrained_weights(trainer.network, args.pretrained_weights)
else:
# new training without pretraine weights, do nothing
pass
trainer.run_training()
else:
if valbest:
trainer.load_best_checkpoint(train=False)
else:
trainer.load_final_checkpoint(train=False)
trainer.network.eval()
if args.disable_validation_inference:
print("Validation inference was disabled. Not running inference on validation set.")
else:
# predict validation
trainer.validate(save_softmax=args.npz, validation_folder_name=val_folder,
run_postprocessing_on_folds=not disable_postprocessing_on_folds,
overwrite=args.val_disable_overwrite)
if network == '3d_lowres' and not args.disable_next_stage_pred:
print("predicting segmentations for the next stage of the cascade")
predict_next_stage(trainer, join(dataset_directory, trainer.plans['data_identifier'] + "_stage%d" % 1))
if __name__ == "__main__":
main()
nnunet/network_architecture/ —— 网络结构核心模块
| 文件 | 作用 |
|---|---|
custom_modules.py |
自定义模块集合,比如特殊卷积、注意力机制、残差连接等,供其他网络复用。 |
initialization.py |
网络权重初始化方法(如 Xavier、He),确保训练稳定。 |
neural_network.py |
基础神经网络类,封装了前向传播、参数管理等通用功能。 |
generic_modular_UNet.py |
基础可配置 UNet 模块化实现,支持任意深度、通道数,是大多数训练器的基础。 |
generic_UNet.py |
经典 UNet 实现(非模块化),较老版本,已逐渐被 generic_modular_UNet 替代。 |
generic_modular_residual_UNet.py |
带残差连接的模块化 UNet,提升梯度流动,适合更深网络。 |
generic_modular_preact_residual_UNet.py |
预激活残差 UNet,即先激活再卷积(pre-activation residual),性能更强,用于高级配置。 |
generic_UNet_DP.py |
支持 数据并行(Data Parallel) 的 UNet 版本,用于多 GPU 训练。 |
__init__.py |
导出模块,让外部可以方便地导入这些网络类。 |
由我的训练命令可以知道我使用的是 generic_modular_residual_UNet.py 中定义的 残差3D UNet,由 nnUNetTrainerV2 自动加载并配置,是 Synapse 任务的标准选择。
export nnUNet_raw_data_base="/xujiheng/Synapse/nnUNet/nnUNet/nnUNetFrame/DATASET/nnUNet_raw" && export nnUNet_preprocessed="/xujiheng/Synapse/nnUNet/nnUNet/nnUNetFrame/DATASET/nnUNet_preprocessed" && export RESULTS_FOLDER="/xujiheng/Synapse/nnUNet/nnUNet/nnUNetFrame/DATASET/nnUNet_trained_models" && python /xujiheng/Synapse/nnUNet/nnUNet/nnunet/run/run_training.py 3d_fullres nnUNetTrainerV2 500 all这是因为 nnUNetTrainerV2 在初始化时主动导入并调用了它来构建默认的残差UNet架构。这是 nnU-Net V2 的标准行为,无需手动指定。
generic_modular_residual_UNet.py:
# Copyright 2020 Division of Medical Image Computing, German Cancer Research Center (DKFZ), Heidelberg, Germany
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import numpy as np
import torch
from nnunet.network_architecture.custom_modules.conv_blocks import BasicResidualBlock, ResidualLayer
from nnunet.network_architecture.generic_UNet import Upsample
from nnunet.network_architecture.generic_modular_UNet import PlainConvUNetDecoder, get_default_network_config
from nnunet.network_architecture.neural_network import SegmentationNetwork
from nnunet.training.loss_functions.dice_loss import DC_and_CE_loss
from torch import nn
from torch.optim import SGD
from torch.backends import cudnn
class ResidualUNetEncoder(nn.Module):
def __init__(self, input_channels, base_num_features, num_blocks_per_stage, feat_map_mul_on_downscale,
pool_op_kernel_sizes, conv_kernel_sizes, props, default_return_skips=True,
max_num_features=480, block=BasicResidualBlock, block_kwargs=None):
"""
Following UNet building blocks can be added by utilizing the properties this class exposes
this one includes the bottleneck layer!
:param input_channels:
:param base_num_features:
:param num_blocks_per_stage:
:param feat_map_mul_on_downscale:
:param pool_op_kernel_sizes:
:param conv_kernel_sizes:
:param props:
"""
super(ResidualUNetEncoder, self).__init__()
if block_kwargs is None:
block_kwargs = {}
self.default_return_skips = default_return_skips
self.props = props
self.stages = []
self.stage_output_features = []
self.stage_pool_kernel_size = []
self.stage_conv_op_kernel_size = []
assert len(pool_op_kernel_sizes) == len(conv_kernel_sizes)
num_stages = len(conv_kernel_sizes)
if not isinstance(num_blocks_per_stage, (list, tuple)):
num_blocks_per_stage = [num_blocks_per_stage] * num_stages
else:
assert len(num_blocks_per_stage) == num_stages
self.num_blocks_per_stage = num_blocks_per_stage # decoder may need this
self.initial_conv = props['conv_op'](input_channels, base_num_features, 3, padding=1, **props['conv_op_kwargs'])
self.initial_norm = props['norm_op'](base_num_features, **props['norm_op_kwargs'])
self.initial_nonlin = props['nonlin'](**props['nonlin_kwargs'])
current_input_features = base_num_features
for stage in range(num_stages):
current_output_features = min(base_num_features * feat_map_mul_on_downscale ** stage, max_num_features)
current_kernel_size = conv_kernel_sizes[stage]
current_pool_kernel_size = pool_op_kernel_sizes[stage]
current_stage = ResidualLayer(current_input_features, current_output_features, current_kernel_size, props,
self.num_blocks_per_stage[stage], current_pool_kernel_size, block,
block_kwargs)
self.stages.append(current_stage)
self.stage_output_features.append(current_stage.output_channels)
self.stage_conv_op_kernel_size.append(current_kernel_size)
self.stage_pool_kernel_size.append(current_pool_kernel_size)
# update current_input_features
current_input_features = current_stage.output_channels
self.output_features = current_input_features
self.stages = nn.ModuleList(self.stages)
def forward(self, x, return_skips=None):
"""
:param x:
:param return_skips: if none then self.default_return_skips is used
:return:
"""
skips = []
x = self.initial_nonlin(self.initial_norm(self.initial_conv(x)))
for s in self.stages:
x = s(x)
if self.default_return_skips:
skips.append(x)
if return_skips is None:
return_skips = self.default_return_skips
# print(x.shape)
if return_skips:
return skips
else:
return x
@staticmethod
def compute_approx_vram_consumption(patch_size, base_num_features, max_num_features,
num_modalities, pool_op_kernel_sizes, num_conv_per_stage_encoder,
feat_map_mul_on_downscale, batch_size):
npool = len(pool_op_kernel_sizes) - 1
current_shape = np.array(patch_size)
tmp = (num_conv_per_stage_encoder[0] * 2 + 1) * np.prod(current_shape) * base_num_features \
+ num_modalities * np.prod(current_shape)
num_feat = base_num_features
for p in range(1, npool + 1):
current_shape = current_shape / np.array(pool_op_kernel_sizes[p])
num_feat = min(num_feat * feat_map_mul_on_downscale, max_num_features)
num_convs = num_conv_per_stage_encoder[p] * 2 + 1 # + 1 for conv in skip in first block
print(p, num_feat, num_convs, current_shape)
tmp += num_convs * np.prod(current_shape) * num_feat
return tmp * batch_size
class ResidualUNetDecoder(nn.Module):
def __init__(self, previous, num_classes, num_blocks_per_stage=None, network_props=None, deep_supervision=False,
upscale_logits=False, block=BasicResidualBlock, block_kwargs=None):
super(ResidualUNetDecoder, self).__init__()
if block_kwargs is None:
block_kwargs = {}
self.num_classes = num_classes
self.deep_supervision = deep_supervision
"""
We assume the bottleneck is part of the encoder, so we can start with upsample -> concat here
"""
previous_stages = previous.stages
previous_stage_output_features = previous.stage_output_features
previous_stage_pool_kernel_size = previous.stage_pool_kernel_size
previous_stage_conv_op_kernel_size = previous.stage_conv_op_kernel_size
if network_props is None:
self.props = previous.props
else:
self.props = network_props
if self.props['conv_op'] == nn.Conv2d:
transpconv = nn.ConvTranspose2d
upsample_mode = "bilinear"
elif self.props['conv_op'] == nn.Conv3d:
transpconv = nn.ConvTranspose3d
upsample_mode = "trilinear"
else:
raise ValueError("unknown convolution dimensionality, conv op: %s" % str(self.props['conv_op']))
if num_blocks_per_stage is None:
num_blocks_per_stage = previous.num_blocks_per_stage[:-1][::-1]
assert len(num_blocks_per_stage) == len(previous.num_blocks_per_stage) - 1
self.stage_pool_kernel_size = previous_stage_pool_kernel_size
self.stage_output_features = previous_stage_output_features
self.stage_conv_op_kernel_size = previous_stage_conv_op_kernel_size
num_stages = len(previous_stages) - 1 # we have one less as the first stage here is what comes after the
# bottleneck
self.tus = []
self.stages = []
self.deep_supervision_outputs = []
# only used for upsample_logits
cum_upsample = np.cumprod(np.vstack(self.stage_pool_kernel_size), axis=0).astype(int)
for i, s in enumerate(np.arange(num_stages)[::-1]):
features_below = previous_stage_output_features[s + 1]
features_skip = previous_stage_output_features[s]
self.tus.append(transpconv(features_below, features_skip, previous_stage_pool_kernel_size[s + 1],
previous_stage_pool_kernel_size[s + 1], bias=False))
# after we tu we concat features so now we have 2xfeatures_skip
self.stages.append(ResidualLayer(2 * features_skip, features_skip, previous_stage_conv_op_kernel_size[s],
self.props, num_blocks_per_stage[i], None, block, block_kwargs))
if deep_supervision and s != 0:
seg_layer = self.props['conv_op'](features_skip, num_classes, 1, 1, 0, 1, 1, bias=True)
if upscale_logits:
upsample = Upsample(scale_factor=cum_upsample[s], mode=upsample_mode)
self.deep_supervision_outputs.append(nn.Sequential(seg_layer, upsample))
else:
self.deep_supervision_outputs.append(seg_layer)
self.segmentation_output = self.props['conv_op'](features_skip, num_classes, 1, 1, 0, 1, 1, bias=True)
self.tus = nn.ModuleList(self.tus)
self.stages = nn.ModuleList(self.stages)
self.deep_supervision_outputs = nn.ModuleList(self.deep_supervision_outputs)
def forward(self, skips):
# skips come from the encoder. They are sorted so that the bottleneck is last in the list
# what is maybe not perfect is that the TUs and stages here are sorted the other way around
# so let's just reverse the order of skips
skips = skips[::-1]
seg_outputs = []
x = skips[0] # this is the bottleneck
for i in range(len(self.tus)):
x = self.tus[i](x)
x = torch.cat((x, skips[i + 1]), dim=1)
x = self.stages[i](x)
if self.deep_supervision and (i != len(self.tus) - 1):
seg_outputs.append(self.deep_supervision_outputs[i](x))
segmentation = self.segmentation_output(x)
if self.deep_supervision:
seg_outputs.append(segmentation)
return seg_outputs[
::-1] # seg_outputs are ordered so that the seg from the highest layer is first, the seg from
# the bottleneck of the UNet last
else:
return segmentation
@staticmethod
def compute_approx_vram_consumption(patch_size, base_num_features, max_num_features,
num_classes, pool_op_kernel_sizes, num_blocks_per_stage_decoder,
feat_map_mul_on_downscale, batch_size):
"""
This only applies for num_conv_per_stage and convolutional_upsampling=True
not real vram consumption. just a constant term to which the vram consumption will be approx proportional
(+ offset for parameter storage)
:param patch_size:
:param num_pool_per_axis:
:param base_num_features:
:param max_num_features:
:return:
"""
npool = len(pool_op_kernel_sizes) - 1
current_shape = np.array(patch_size)
tmp = (num_blocks_per_stage_decoder[-1] * 2 + 1) * np.prod(
current_shape) * base_num_features + num_classes * np.prod(current_shape)
num_feat = base_num_features
for p in range(1, npool):
current_shape = current_shape / np.array(pool_op_kernel_sizes[p])
num_feat = min(num_feat * feat_map_mul_on_downscale, max_num_features)
num_convs = num_blocks_per_stage_decoder[-(p + 1)] * 2 + 1 + 1 # +1 for transpconv and +1 for conv in skip
print(p, num_feat, num_convs, current_shape)
tmp += num_convs * np.prod(current_shape) * num_feat
return tmp * batch_size
class ResidualUNet(SegmentationNetwork):
use_this_for_batch_size_computation_2D = 858931200.0 # 1167982592.0
use_this_for_batch_size_computation_3D = 727842816.0 # 1152286720.0
default_base_num_features = 24
default_conv_per_stage = (2, 2, 2, 2, 2, 2, 2, 2)
def __init__(self, input_channels, base_num_features, num_blocks_per_stage_encoder, feat_map_mul_on_downscale,
pool_op_kernel_sizes, conv_kernel_sizes, props, num_classes, num_blocks_per_stage_decoder,
deep_supervision=False, upscale_logits=False, max_features=512, initializer=None,
block=BasicResidualBlock, block_kwargs=None):
super(ResidualUNet, self).__init__()
if block_kwargs is None:
block_kwargs = {}
self.conv_op = props['conv_op']
self.num_classes = num_classes
self.encoder = ResidualUNetEncoder(input_channels, base_num_features, num_blocks_per_stage_encoder,
feat_map_mul_on_downscale, pool_op_kernel_sizes, conv_kernel_sizes,
props, default_return_skips=True, max_num_features=max_features,
block=block, block_kwargs=block_kwargs)
self.decoder = ResidualUNetDecoder(self.encoder, num_classes, num_blocks_per_stage_decoder, props,
deep_supervision, upscale_logits, block=block, block_kwargs=block_kwargs)
if initializer is not None:
self.apply(initializer)
def forward(self, x):
skips = self.encoder(x)
return self.decoder(skips)
@staticmethod
def compute_approx_vram_consumption(patch_size, base_num_features, max_num_features,
num_modalities, num_classes, pool_op_kernel_sizes, num_conv_per_stage_encoder,
num_conv_per_stage_decoder, feat_map_mul_on_downscale, batch_size):
enc = ResidualUNetEncoder.compute_approx_vram_consumption(patch_size, base_num_features, max_num_features,
num_modalities, pool_op_kernel_sizes,
num_conv_per_stage_encoder,
feat_map_mul_on_downscale, batch_size)
dec = ResidualUNetDecoder.compute_approx_vram_consumption(patch_size, base_num_features, max_num_features,
num_classes, pool_op_kernel_sizes,
num_conv_per_stage_decoder,
feat_map_mul_on_downscale, batch_size)
return enc + dec
class FabiansUNet(SegmentationNetwork):
"""
Residual Encoder, Plain conv decoder
"""
use_this_for_2D_configuration = 1244233721.0 # 1167982592.0
use_this_for_3D_configuration = 1230348801.0
default_blocks_per_stage_encoder = (1, 2, 3, 4, 4, 4, 4, 4, 4, 4, 4)
default_blocks_per_stage_decoder = (1, 1, 1, 1, 1, 1, 1, 1, 1, 1)
default_min_batch_size = 2 # this is what works with the numbers above
def __init__(self, input_channels, base_num_features, num_blocks_per_stage_encoder, feat_map_mul_on_downscale,
pool_op_kernel_sizes, conv_kernel_sizes, props, num_classes, num_blocks_per_stage_decoder,
deep_supervision=False, upscale_logits=False, max_features=512, initializer=None,
block=BasicResidualBlock,
props_decoder=None, block_kwargs=None):
super().__init__()
if block_kwargs is None:
block_kwargs = {}
self.conv_op = props['conv_op']
self.num_classes = num_classes
self.encoder = ResidualUNetEncoder(input_channels, base_num_features, num_blocks_per_stage_encoder,
feat_map_mul_on_downscale, pool_op_kernel_sizes, conv_kernel_sizes,
props, default_return_skips=True, max_num_features=max_features,
block=block, block_kwargs=block_kwargs)
props['dropout_op_kwargs']['p'] = 0
if props_decoder is None:
props_decoder = props
self.decoder = PlainConvUNetDecoder(self.encoder, num_classes, num_blocks_per_stage_decoder, props_decoder,
deep_supervision, upscale_logits)
if initializer is not None:
self.apply(initializer)
def forward(self, x):
skips = self.encoder(x)
return self.decoder(skips)
@staticmethod
def compute_approx_vram_consumption(patch_size, base_num_features, max_num_features,
num_modalities, num_classes, pool_op_kernel_sizes, num_conv_per_stage_encoder,
num_conv_per_stage_decoder, feat_map_mul_on_downscale, batch_size):
enc = ResidualUNetEncoder.compute_approx_vram_consumption(patch_size, base_num_features, max_num_features,
num_modalities, pool_op_kernel_sizes,
num_conv_per_stage_encoder,
feat_map_mul_on_downscale, batch_size)
dec = PlainConvUNetDecoder.compute_approx_vram_consumption(patch_size, base_num_features, max_num_features,
num_classes, pool_op_kernel_sizes,
num_conv_per_stage_decoder,
feat_map_mul_on_downscale, batch_size)
return enc + dec
def find_3d_configuration():
# lets compute a reference for 3D
# we select hyperparameters here so that we get approximately the same patch size as we would get with the
# regular unet. This is just my choice. You can do whatever you want
# These default hyperparemeters will then be used by the experiment planner
# since this is more parameter intensive than the UNet, we will test a configuration that has a lot of parameters
# herefore we copy the UNet configuration for Task005_Prostate
cudnn.deterministic = False
cudnn.benchmark = True
patch_size = (20, 320, 256)
max_num_features = 320
num_modalities = 2
num_classes = 3
batch_size = 2
# now we fiddle with the network specific hyperparameters until everything just barely fits into a titanx
blocks_per_stage_encoder = FabiansUNet.default_blocks_per_stage_encoder
blocks_per_stage_decoder = FabiansUNet.default_blocks_per_stage_decoder
initial_num_features = 32
# we neeed to add a [1, 1, 1] for the res unet because in this implementation all stages of the encoder can have a stride
pool_op_kernel_sizes = [[1, 1, 1],
[1, 2, 2],
[1, 2, 2],
[2, 2, 2],
[2, 2, 2],
[1, 2, 2],
[1, 2, 2]]
conv_op_kernel_sizes = [[1, 3, 3],
[1, 3, 3],
[3, 3, 3],
[3, 3, 3],
[3, 3, 3],
[3, 3, 3],
[3, 3, 3]]
unet = FabiansUNet(num_modalities, initial_num_features, blocks_per_stage_encoder[:len(conv_op_kernel_sizes)], 2,
pool_op_kernel_sizes, conv_op_kernel_sizes,
get_default_network_config(3, dropout_p=None), num_classes,
blocks_per_stage_decoder[:len(conv_op_kernel_sizes)-1], False, False,
max_features=max_num_features).cuda()
optimizer = SGD(unet.parameters(), lr=0.1, momentum=0.95)
loss = DC_and_CE_loss({'batch_dice': True, 'smooth': 1e-5, 'do_bg': False}, {})
dummy_input = torch.rand((batch_size, num_modalities, *patch_size)).cuda()
dummy_gt = (torch.rand((batch_size, 1, *patch_size)) * num_classes).round().clamp_(0, 2).cuda().long()
for _ in range(20):
optimizer.zero_grad()
skips = unet.encoder(dummy_input)
print([i.shape for i in skips])
output = unet.decoder(skips)
l = loss(output, dummy_gt)
l.backward()
optimizer.step()
if _ == 0:
torch.cuda.empty_cache()
# that should do. Now take the network hyperparameters and insert them in FabiansUNet.compute_approx_vram_consumption
# whatever number this spits out, save it to FabiansUNet.use_this_for_batch_size_computation_3D
print(FabiansUNet.compute_approx_vram_consumption(patch_size, initial_num_features, max_num_features, num_modalities,
num_classes, pool_op_kernel_sizes,
blocks_per_stage_encoder[:len(conv_op_kernel_sizes)],
blocks_per_stage_decoder[:len(conv_op_kernel_sizes)-1], 2, batch_size))
# the output is 1230348800.0 for me
# I increment that number by 1 to allow this configuration be be chosen
def find_2d_configuration():
# lets compute a reference for 3D
# we select hyperparameters here so that we get approximately the same patch size as we would get with the
# regular unet. This is just my choice. You can do whatever you want
# These default hyperparemeters will then be used by the experiment planner
# since this is more parameter intensive than the UNet, we will test a configuration that has a lot of parameters
# herefore we copy the UNet configuration for Task003_Liver
cudnn.deterministic = False
cudnn.benchmark = True
patch_size = (512, 512)
max_num_features = 512
num_modalities = 1
num_classes = 3
batch_size = 12
# now we fiddle with the network specific hyperparameters until everything just barely fits into a titanx
blocks_per_stage_encoder = FabiansUNet.default_blocks_per_stage_encoder
blocks_per_stage_decoder = FabiansUNet.default_blocks_per_stage_decoder
initial_num_features = 30
# we neeed to add a [1, 1, 1] for the res unet because in this implementation all stages of the encoder can have a stride
pool_op_kernel_sizes = [[1, 1],
[2, 2],
[2, 2],
[2, 2],
[2, 2],
[2, 2],
[2, 2],
[2, 2]]
conv_op_kernel_sizes = [[3, 3],
[3, 3],
[3, 3],
[3, 3],
[3, 3],
[3, 3],
[3, 3],
[3, 3]]
unet = FabiansUNet(num_modalities, initial_num_features, blocks_per_stage_encoder[:len(conv_op_kernel_sizes)], 2,
pool_op_kernel_sizes, conv_op_kernel_sizes,
get_default_network_config(2, dropout_p=None), num_classes,
blocks_per_stage_decoder[:len(conv_op_kernel_sizes)-1], False, False,
max_features=max_num_features).cuda()
optimizer = SGD(unet.parameters(), lr=0.1, momentum=0.95)
loss = DC_and_CE_loss({'batch_dice': True, 'smooth': 1e-5, 'do_bg': False}, {})
dummy_input = torch.rand((batch_size, num_modalities, *patch_size)).cuda()
dummy_gt = (torch.rand((batch_size, 1, *patch_size)) * num_classes).round().clamp_(0, 2).cuda().long()
for _ in range(20):
optimizer.zero_grad()
skips = unet.encoder(dummy_input)
print([i.shape for i in skips])
output = unet.decoder(skips)
l = loss(output, dummy_gt)
l.backward()
optimizer.step()
if _ == 0:
torch.cuda.empty_cache()
# that should do. Now take the network hyperparameters and insert them in FabiansUNet.compute_approx_vram_consumption
# whatever number this spits out, save it to FabiansUNet.use_this_for_batch_size_computation_2D
print(FabiansUNet.compute_approx_vram_consumption(patch_size, initial_num_features, max_num_features, num_modalities,
num_classes, pool_op_kernel_sizes,
blocks_per_stage_encoder[:len(conv_op_kernel_sizes)],
blocks_per_stage_decoder[:len(conv_op_kernel_sizes)-1], 2, batch_size))
# the output is 1244233728.0 for me
# I increment that number by 1 to allow this configuration be be chosen
# This will not fit with 32 filters, but so will the regular U-net. We still use 32 filters in training.
# This does not matter because we are using mixed precision training now, so a rough memory approximation is OK
if __name__ == "__main__":
pass
nnunet/training/network_training/——训练器(Trainer)系统
| 文件 | 作用 |
|---|---|
nnUNetTrainer.py |
最原始的基础训练器,功能简单,已基本不用。 |
nnUNetTrainerV2.py |
✅ 主流默认训练器,支持混合精度、动态学习率、批量 Dice 损失等。 |
nnUNetTrainerV2_DP.py |
支持 数据并行(Data Parallel),用于单机多 GPU 训练。 |
nnUNetTrainerV2_fp32.py |
禁用混合精度,强制使用 FP32,适合调试或兼容性问题。 |
nnUNetTrainerV2_DDP.py |
支持 分布式数据并行(Distributed Data Parallel),用于多机或多卡训练(如 8xGPU)。 |
nnUNetTrainerCascadeFullRes.py |
用于 级联(cascade)模型的全分辨率阶段,输入是低分辨率预测结果。 |
nnUNetTrainerV2_CascadeFullRes.py |
✅ V2 版本的级联全分辨率训练器,现代推荐版本。 |
nnUNet_variants/ |
存放各种实验性或定制化的训练器变体(如注意力机制、不同损失函数等)。 |
competitions_with_custom_Trainers/ |
为特定比赛准备的自定义训练器,一般用户无需关心。 |
nnUNetTrainerV2.py是标准配置,适用于大多数任务:
# Copyright 2020 Division of Medical Image Computing, German Cancer Research Center (DKFZ), Heidelberg, Germany
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
from collections import OrderedDict
from typing import Tuple
import numpy as np
import torch
from nnunet.training.data_augmentation.data_augmentation_moreDA import get_moreDA_augmentation
from nnunet.training.loss_functions.deep_supervision import MultipleOutputLoss2
from nnunet.utilities.to_torch import maybe_to_torch, to_cuda
from nnunet.network_architecture.generic_UNet import Generic_UNet
from nnunet.network_architecture.initialization import InitWeights_He
from nnunet.network_architecture.neural_network import SegmentationNetwork
from nnunet.training.data_augmentation.default_data_augmentation import default_2D_augmentation_params, \
get_patch_size, default_3D_augmentation_params
from nnunet.training.dataloading.dataset_loading import unpack_dataset
from nnunet.training.network_training.nnUNetTrainer import nnUNetTrainer
from nnunet.utilities.nd_softmax import softmax_helper
from sklearn.model_selection import KFold
from torch import nn
from torch.cuda.amp import autocast
from nnunet.training.learning_rate.poly_lr import poly_lr
from batchgenerators.utilities.file_and_folder_operations import *
class nnUNetTrainerV2(nnUNetTrainer):
"""
Info for Fabian: same as internal nnUNetTrainerV2_2
"""
def __init__(self, plans_file, fold, output_folder=None, dataset_directory=None, batch_dice=True, stage=None,
unpack_data=True, deterministic=True, fp16=False):
super().__init__(plans_file, fold, output_folder, dataset_directory, batch_dice, stage, unpack_data,
deterministic, fp16)
self.max_num_epochs = 1000
self.initial_lr = 1e-2
self.deep_supervision_scales = None
self.ds_loss_weights = None
self.pin_memory = True
def initialize(self, training=True, force_load_plans=False):
"""
- replaced get_default_augmentation with get_moreDA_augmentation
- enforce to only run this code once
- loss function wrapper for deep supervision
:param training:
:param force_load_plans:
:return:
"""
if not self.was_initialized:
maybe_mkdir_p(self.output_folder)
if force_load_plans or (self.plans is None):
self.load_plans_file()
self.process_plans(self.plans)
self.setup_DA_params()
################# Here we wrap the loss for deep supervision ############
# we need to know the number of outputs of the network
net_numpool = len(self.net_num_pool_op_kernel_sizes)
# we give each output a weight which decreases exponentially (division by 2) as the resolution decreases
# this gives higher resolution outputs more weight in the loss
weights = np.array([1 / (2 ** i) for i in range(net_numpool)])
# we don't use the lowest 2 outputs. Normalize weights so that they sum to 1
mask = np.array([True] + [True if i < net_numpool - 1 else False for i in range(1, net_numpool)])
weights[~mask] = 0
weights = weights / weights.sum()
self.ds_loss_weights = weights
# now wrap the loss
self.loss = MultipleOutputLoss2(self.loss, self.ds_loss_weights)
################# END ###################
self.folder_with_preprocessed_data = join(self.dataset_directory, self.plans['data_identifier'] +
"_stage%d" % self.stage)
if training:
self.dl_tr, self.dl_val = self.get_basic_generators()
if self.unpack_data:
print("unpacking dataset")
unpack_dataset(self.folder_with_preprocessed_data)
print("done")
else:
print(
"INFO: Not unpacking data! Training may be slow due to that. Pray you are not using 2d or you "
"will wait all winter for your model to finish!")
self.tr_gen, self.val_gen = get_moreDA_augmentation(
self.dl_tr, self.dl_val,
self.data_aug_params[
'patch_size_for_spatialtransform'],
self.data_aug_params,
deep_supervision_scales=self.deep_supervision_scales,
pin_memory=self.pin_memory,
use_nondetMultiThreadedAugmenter=False
)
self.print_to_log_file("TRAINING KEYS:\n %s" % (str(self.dataset_tr.keys())),
also_print_to_console=False)
self.print_to_log_file("VALIDATION KEYS:\n %s" % (str(self.dataset_val.keys())),
also_print_to_console=False)
else:
pass
self.initialize_network()
self.initialize_optimizer_and_scheduler()
assert isinstance(self.network, (SegmentationNetwork, nn.DataParallel))
else:
self.print_to_log_file('self.was_initialized is True, not running self.initialize again')
self.was_initialized = True
def initialize_network(self):
"""
- momentum 0.99
- SGD instead of Adam
- self.lr_scheduler = None because we do poly_lr
- deep supervision = True
- i am sure I forgot something here
Known issue: forgot to set neg_slope=0 in InitWeights_He; should not make a difference though
:return:
"""
if self.threeD:
conv_op = nn.Conv3d
dropout_op = nn.Dropout3d
norm_op = nn.InstanceNorm3d
else:
conv_op = nn.Conv2d
dropout_op = nn.Dropout2d
norm_op = nn.InstanceNorm2d
norm_op_kwargs = {'eps': 1e-5, 'affine': True}
dropout_op_kwargs = {'p': 0, 'inplace': True}
net_nonlin = nn.LeakyReLU
net_nonlin_kwargs = {'negative_slope': 1e-2, 'inplace': True}
self.network = Generic_UNet(self.num_input_channels, self.base_num_features, self.num_classes,
len(self.net_num_pool_op_kernel_sizes),
self.conv_per_stage, 2, conv_op, norm_op, norm_op_kwargs, dropout_op,
dropout_op_kwargs,
net_nonlin, net_nonlin_kwargs, True, False, lambda x: x, InitWeights_He(1e-2),
self.net_num_pool_op_kernel_sizes, self.net_conv_kernel_sizes, False, True, True)
if torch.cuda.is_available():
self.network.cuda()
self.network.inference_apply_nonlin = softmax_helper
def initialize_optimizer_and_scheduler(self):
assert self.network is not None, "self.initialize_network must be called first"
self.optimizer = torch.optim.SGD(self.network.parameters(), self.initial_lr, weight_decay=self.weight_decay,
momentum=0.99, nesterov=True)
self.lr_scheduler = None
def run_online_evaluation(self, output, target):
"""
due to deep supervision the return value and the reference are now lists of tensors. We only need the full
resolution output because this is what we are interested in in the end. The others are ignored
:param output:
:param target:
:return:
"""
target = target[0]
output = output[0]
return super().run_online_evaluation(output, target)
def validate(self, do_mirroring: bool = True, use_sliding_window: bool = True,
step_size: float = 0.5, save_softmax: bool = True, use_gaussian: bool = True, overwrite: bool = True,
validation_folder_name: str = 'validation_raw', debug: bool = False, all_in_gpu: bool = False,
segmentation_export_kwargs: dict = None, run_postprocessing_on_folds: bool = True):
"""
We need to wrap this because we need to enforce self.network.do_ds = False for prediction
"""
ds = self.network.do_ds
self.network.do_ds = False
ret = super().validate(do_mirroring=do_mirroring, use_sliding_window=use_sliding_window, step_size=step_size,
save_softmax=save_softmax, use_gaussian=use_gaussian,
overwrite=overwrite, validation_folder_name=validation_folder_name, debug=debug,
all_in_gpu=all_in_gpu, segmentation_export_kwargs=segmentation_export_kwargs,
run_postprocessing_on_folds=run_postprocessing_on_folds)
self.network.do_ds = ds
return ret
def predict_preprocessed_data_return_seg_and_softmax(self, data: np.ndarray, do_mirroring: bool = True,
mirror_axes: Tuple[int] = None,
use_sliding_window: bool = True, step_size: float = 0.5,
use_gaussian: bool = True, pad_border_mode: str = 'constant',
pad_kwargs: dict = None, all_in_gpu: bool = False,
verbose: bool = True, mixed_precision=True) -> Tuple[np.ndarray, np.ndarray]:
"""
We need to wrap this because we need to enforce self.network.do_ds = False for prediction
"""
ds = self.network.do_ds
self.network.do_ds = False
ret = super().predict_preprocessed_data_return_seg_and_softmax(data,
do_mirroring=do_mirroring,
mirror_axes=mirror_axes,
use_sliding_window=use_sliding_window,
step_size=step_size, use_gaussian=use_gaussian,
pad_border_mode=pad_border_mode,
pad_kwargs=pad_kwargs, all_in_gpu=all_in_gpu,
verbose=verbose,
mixed_precision=mixed_precision)
self.network.do_ds = ds
return ret
def run_iteration(self, data_generator, do_backprop=True, run_online_evaluation=False):
"""
gradient clipping improves training stability
:param data_generator:
:param do_backprop:
:param run_online_evaluation:
:return:
"""
data_dict = next(data_generator)
data = data_dict['data']
target = data_dict['target']
data = maybe_to_torch(data)
target = maybe_to_torch(target)
if torch.cuda.is_available():
data = to_cuda(data)
target = to_cuda(target)
self.optimizer.zero_grad()
if self.fp16:
with autocast():
output = self.network(data)
del data
l = self.loss(output, target)
if do_backprop:
self.amp_grad_scaler.scale(l).backward()
self.amp_grad_scaler.unscale_(self.optimizer)
torch.nn.utils.clip_grad_norm_(self.network.parameters(), 12)
self.amp_grad_scaler.step(self.optimizer)
self.amp_grad_scaler.update()
else:
output = self.network(data)
del data
l = self.loss(output, target)
if do_backprop:
l.backward()
torch.nn.utils.clip_grad_norm_(self.network.parameters(), 12)
self.optimizer.step()
if run_online_evaluation:
self.run_online_evaluation(output, target)
del target
return l.detach().cpu().numpy()
def do_split(self):
"""
The default split is a 5 fold CV on all available training cases. nnU-Net will create a split (it is seeded,
so always the same) and save it as splits_final.pkl file in the preprocessed data directory.
Sometimes you may want to create your own split for various reasons. For this you will need to create your own
splits_final.pkl file. If this file is present, nnU-Net is going to use it and whatever splits are defined in
it. You can create as many splits in this file as you want. Note that if you define only 4 splits (fold 0-3)
and then set fold=4 when training (that would be the fifth split), nnU-Net will print a warning and proceed to
use a random 80:20 data split.
:return:
"""
if self.fold == "all":
# if fold==all then we use all images for training and validation
tr_keys = val_keys = list(self.dataset.keys())
else:
splits_file = join(self.dataset_directory, "splits_final.pkl")
# if the split file does not exist we need to create it
if not isfile(splits_file):
self.print_to_log_file("Creating new 5-fold cross-validation split...")
splits = []
all_keys_sorted = np.sort(list(self.dataset.keys()))
kfold = KFold(n_splits=5, shuffle=True, random_state=12345)
for i, (train_idx, test_idx) in enumerate(kfold.split(all_keys_sorted)):
train_keys = np.array(all_keys_sorted)[train_idx]
test_keys = np.array(all_keys_sorted)[test_idx]
splits.append(OrderedDict())
splits[-1]['train'] = train_keys
splits[-1]['val'] = test_keys
save_pickle(splits, splits_file)
else:
self.print_to_log_file("Using splits from existing split file:", splits_file)
splits = load_pickle(splits_file)
self.print_to_log_file("The split file contains %d splits." % len(splits))
self.print_to_log_file("Desired fold for training: %d" % self.fold)
if self.fold < len(splits):
tr_keys = splits[self.fold]['train']
val_keys = splits[self.fold]['val']
self.print_to_log_file("This split has %d training and %d validation cases."
% (len(tr_keys), len(val_keys)))
else:
self.print_to_log_file("INFO: You requested fold %d for training but splits "
"contain only %d folds. I am now creating a "
"random (but seeded) 80:20 split!" % (self.fold, len(splits)))
# if we request a fold that is not in the split file, create a random 80:20 split
rnd = np.random.RandomState(seed=12345 + self.fold)
keys = np.sort(list(self.dataset.keys()))
idx_tr = rnd.choice(len(keys), int(len(keys) * 0.8), replace=False)
idx_val = [i for i in range(len(keys)) if i not in idx_tr]
tr_keys = [keys[i] for i in idx_tr]
val_keys = [keys[i] for i in idx_val]
self.print_to_log_file("This random 80:20 split has %d training and %d validation cases."
% (len(tr_keys), len(val_keys)))
tr_keys.sort()
val_keys.sort()
self.dataset_tr = OrderedDict()
for i in tr_keys:
self.dataset_tr[i] = self.dataset[i]
self.dataset_val = OrderedDict()
for i in val_keys:
self.dataset_val[i] = self.dataset[i]
def setup_DA_params(self):
"""
- we increase roation angle from [-15, 15] to [-30, 30]
- scale range is now (0.7, 1.4), was (0.85, 1.25)
- we don't do elastic deformation anymore
:return:
"""
self.deep_supervision_scales = [[1, 1, 1]] + list(list(i) for i in 1 / np.cumprod(
np.vstack(self.net_num_pool_op_kernel_sizes), axis=0))[:-1]
if self.threeD:
self.data_aug_params = default_3D_augmentation_params
self.data_aug_params['rotation_x'] = (-30. / 360 * 2. * np.pi, 30. / 360 * 2. * np.pi)
self.data_aug_params['rotation_y'] = (-30. / 360 * 2. * np.pi, 30. / 360 * 2. * np.pi)
self.data_aug_params['rotation_z'] = (-30. / 360 * 2. * np.pi, 30. / 360 * 2. * np.pi)
if self.do_dummy_2D_aug:
self.data_aug_params["dummy_2D"] = True
self.print_to_log_file("Using dummy2d data augmentation")
self.data_aug_params["elastic_deform_alpha"] = \
default_2D_augmentation_params["elastic_deform_alpha"]
self.data_aug_params["elastic_deform_sigma"] = \
default_2D_augmentation_params["elastic_deform_sigma"]
self.data_aug_params["rotation_x"] = default_2D_augmentation_params["rotation_x"]
else:
self.do_dummy_2D_aug = False
if max(self.patch_size) / min(self.patch_size) > 1.5:
default_2D_augmentation_params['rotation_x'] = (-15. / 360 * 2. * np.pi, 15. / 360 * 2. * np.pi)
self.data_aug_params = default_2D_augmentation_params
self.data_aug_params["mask_was_used_for_normalization"] = self.use_mask_for_norm
if self.do_dummy_2D_aug:
self.basic_generator_patch_size = get_patch_size(self.patch_size[1:],
self.data_aug_params['rotation_x'],
self.data_aug_params['rotation_y'],
self.data_aug_params['rotation_z'],
self.data_aug_params['scale_range'])
self.basic_generator_patch_size = np.array([self.patch_size[0]] + list(self.basic_generator_patch_size))
else:
self.basic_generator_patch_size = get_patch_size(self.patch_size, self.data_aug_params['rotation_x'],
self.data_aug_params['rotation_y'],
self.data_aug_params['rotation_z'],
self.data_aug_params['scale_range'])
self.data_aug_params["scale_range"] = (0.7, 1.4)
self.data_aug_params["do_elastic"] = False
self.data_aug_params['selected_seg_channels'] = [0]
self.data_aug_params['patch_size_for_spatialtransform'] = self.patch_size
self.data_aug_params["num_cached_per_thread"] = 2
def maybe_update_lr(self, epoch=None):
"""
if epoch is not None we overwrite epoch. Else we use epoch = self.epoch + 1
(maybe_update_lr is called in on_epoch_end which is called before epoch is incremented.
herefore we need to do +1 here)
:param epoch:
:return:
"""
if epoch is None:
ep = self.epoch + 1
else:
ep = epoch
self.optimizer.param_groups[0]['lr'] = poly_lr(ep, self.max_num_epochs, self.initial_lr, 0.9)
self.print_to_log_file("lr:", np.round(self.optimizer.param_groups[0]['lr'], decimals=6))
def on_epoch_end(self):
"""
overwrite patient-based early stopping. Always run to 1000 epochs
:return:
"""
super().on_epoch_end()
continue_training = self.epoch < self.max_num_epochs
# it can rarely happen that the momentum of nnUNetTrainerV2 is too high for some dataset. If at epoch 100 the
# estimated validation Dice is still 0 then we reduce the momentum from 0.99 to 0.95
if self.epoch == 100:
if self.all_val_eval_metrics[-1] == 0:
self.optimizer.param_groups[0]["momentum"] = 0.95
self.network.apply(InitWeights_He(1e-2))
self.print_to_log_file("At epoch 100, the mean foreground Dice was 0. This can be caused by a too "
"high momentum. High momentum (0.99) is good for datasets where it works, but "
"sometimes causes issues such as this one. Momentum has now been reduced to "
"0.95 and network weights have been reinitialized")
return continue_training
def run_training(self):
"""
if we run with -c then we need to set the correct lr for the first epoch, otherwise it will run the first
continued epoch with self.initial_lr
we also need to make sure deep supervision in the network is enabled for training, thus the wrapper
:return:
"""
self.maybe_update_lr(self.epoch) # if we dont overwrite epoch then self.epoch+1 is used which is not what we
# want at the start of the training
ds = self.network.do_ds
self.network.do_ds = True
ret = super().run_training()
self.network.do_ds = ds
return ret
nnUNetTrainerV2 会按以下流程工作:
(1)初始化阶段(__init__ + initialize())
读取计划文件:从 plans.pkl 中获取:
输入通道数(如 1 for CT)
类别数(如 5 for organs)
网络池化层数、卷积核大小
是否 3D
设置超参数:
最大训练轮数:max_num_epochs = 1000
初始学习率:initial_lr = 0.01
使用 SGD + 动量 0.99 + Nesterov
配置数据增强策略:
旋转 ±30°,缩放 0.7~1.4 倍
不用弹性形变
准备深层监督:
计算每个尺度输出的损失权重(越高层权重越低)
用 MultipleOutputLoss2 包装损失函数
(2)构建网络(initialize_network())
实例化 Generic_UNet 网络
根据 2D/3D 选择合适的卷积层、归一化层
设置激活函数为 LeakyReLU(0.01)
权重初始化(He 初始化)
把网络搬到 GPU 上
设置推理时使用 softmax
(3)准备数据(do_split() + get_basic_generators())
读取或生成 5 折交叉验证划分(splits_final.pkl)
分出训练集 (dataset_tr) 和验证集 (dataset_val)
创建数据加载器 dl_tr, dl_val
使用 get_moreDA_augmentation 添加实时数据增强
每次送入网络前都随机旋转、缩放、翻转等
(4)开始训练(run_training() → run_iteration())
进入主循环,每一轮(epoch)训练一个 epoch:
支持混合精度训练(FP16)→ 更快、更省显存
梯度裁剪(clip_grad_norm_)→ 防止梯度爆炸
每个 epoch 结束后更新学习率(poly_lr)
验证一个 epoch:
关闭深层监督(只保留最终输出)
在验证集上预测
计算 Dice 分数
保存当前最优模型(model_best.model)
(5)收尾工作(训练结束)
保存最终模型(model_final_checkpoint.model)
保存训练日志(training_log.json)
nnunet/inference/——推理与评估
| 文件/文件夹名 | 类型 | 功能简介 | 是否核心 | 使用场景 |
|---|---|---|---|---|
predict.py |
脚本 | 官方主推理脚本,支持滑动窗口、多GPU、batch推理、自动加载模型与后处理。功能完整,推荐用于正式项目。 | 是 | 所有推理任务的首选 |
predict_simple.py |
脚本 | 简化版推理脚本,适合快速测试或调试单个样本,功能较基础,不支持多GPU或集成。 | 是 | 初学者快速验证模型 |
pretrained_models/ |
目录 | 存放预训练模型(如公开挑战赛模型),结构与 nnUNet_trained_models 一致,可通过 -p 参数指定路径加载。 |
可选 | 使用第三方模型时 |
ensemble_predictions.py |
脚本 | 实现多模型集成预测(如5折交叉验证的fold结果融合),提升分割稳定性与性能,常用于比赛或论文实验。 | 是 | 模型融合、性能优化 |
__init__.py |
文件 | Python 包初始化文件,导出推理接口(如 predict 函数),使该目录可被导入使用。 |
是 | 内部机制,无需手动修改 |
change_trainer.py |
脚本 | 在推理时动态更换 Trainer 类,用于比较不同网络结构(如 Unet vs SwinUNet)的表现,无需重新训练。 | 是 | 模型对比实验 |
segmentation_export.py |
脚本 | 将预测结果导出为临床可用格式(如 DICOM、NIfTI + JSON),适用于部署到医院系统或PACS。 | 是 | 模型落地与部署 |
amos2022/ |
目录 | 针对 AMOS 2022 挑战赛 的定制化配置,可能包含特定标签映射、预处理或后处理逻辑。 | 否 | 仅限 AMOS 任务使用 |
__pycache__/ |
目录 | Python 自动生成的缓存文件夹(.pyc),加速模块导入,可安全删除,不影响功能。 |
否 | 系统自动生成,无需关注 |
这里是训练后输出的所有结果:
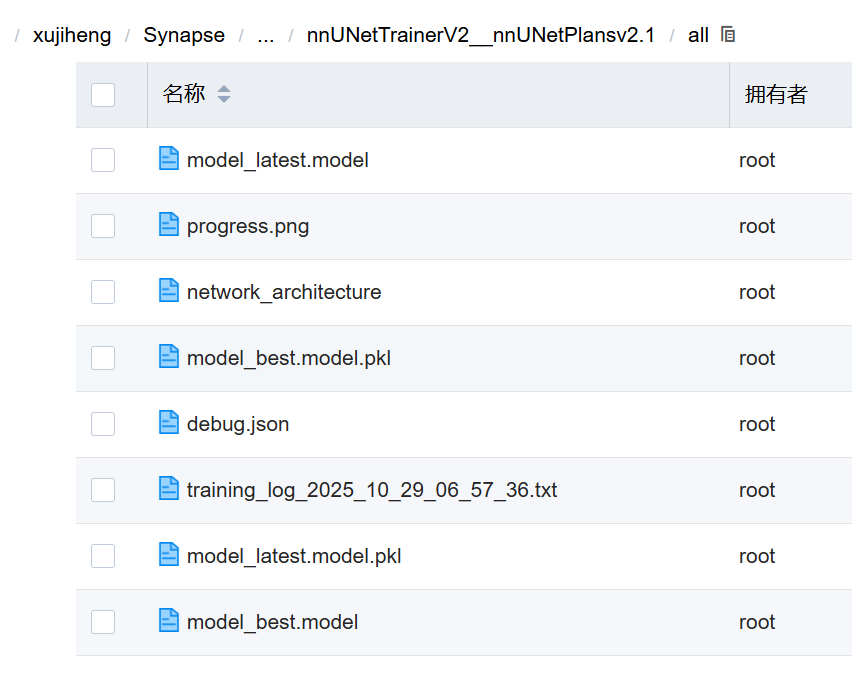
| 文件名 | 类型 | 功能 | 是否核心 |
|---|---|---|---|
model_best.model |
模型权重 | 训练过程中 Dice 最高 的模型检查点(推荐用于推理) | 是 |
model_best.model.pkl |
网络结构 | 包含网络架构信息(如 layer_num, kernel_size),与 .model 配对使用 |
是 |
model_latest.model |
模型权重 | 最后一个 epoch 的模型(不一定是性能最好的) | 可选 |
model_latest.model.pkl |
网络结构 | 对应 model_latest.model 的结构文件 |
可选 |
progress.png |
图像 | 训练过程中的 loss 和 Dice 曲线图(直观查看收敛情况) | 推荐查看 |
network_architecture |
文本 | 网络结构的文本描述(如 UNet, depth=4, filters=32) | 了解模型配置 |
debug.json |
JSON | 调试信息(如 batch size、patch size、optimizer 参数) | 重要,用于复现 |
training_log_...txt |
日志 | 完整训练日志(每轮 loss、Dice、时间等) | 必看 |
predict.py:
# Copyright 2020 Division of Medical Image Computing, German Cancer Research Center (DKFZ), Heidelberg, Germany
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import argparse
from copy import deepcopy
from typing import Tuple, Union, List
import numpy as np
from batchgenerators.augmentations.utils import resize_segmentation
from nnunet.inference.segmentation_export import save_segmentation_nifti_from_softmax, save_segmentation_nifti
from batchgenerators.utilities.file_and_folder_operations import *
import sys
if 'win' in sys.platform:
#fix for windows platform
import pathos
Process = pathos.helpers.mp.Process
Queue = pathos.helpers.mp.Queue
else:
from multiprocessing import Process, Queue
import torch
import SimpleITK as sitk
import shutil
from multiprocessing import Pool
from nnunet.postprocessing.connected_components import load_remove_save, load_postprocessing
from nnunet.training.model_restore import load_model_and_checkpoint_files
from nnunet.training.network_training.nnUNetTrainer import nnUNetTrainer
from nnunet.utilities.one_hot_encoding import to_one_hot
def preprocess_save_to_queue(preprocess_fn, q, list_of_lists, output_files, segs_from_prev_stage, classes,
transpose_forward):
# suppress output
# sys.stdout = open(os.devnull, 'w')
errors_in = []
for i, l in enumerate(list_of_lists):
try:
output_file = output_files[i]
print("preprocessing", output_file)
d, _, dct = preprocess_fn(l)
# print(output_file, dct)
if segs_from_prev_stage[i] is not None:
assert isfile(segs_from_prev_stage[i]) and segs_from_prev_stage[i].endswith(
".nii.gz"), "segs_from_prev_stage" \
" must point to a " \
"segmentation file"
seg_prev = sitk.GetArrayFromImage(sitk.ReadImage(segs_from_prev_stage[i]))
# check to see if shapes match
img = sitk.GetArrayFromImage(sitk.ReadImage(l[0]))
assert all([i == j for i, j in zip(seg_prev.shape, img.shape)]), "image and segmentation from previous " \
"stage don't have the same pixel array " \
"shape! image: %s, seg_prev: %s" % \
(l[0], segs_from_prev_stage[i])
seg_prev = seg_prev.transpose(transpose_forward)
seg_reshaped = resize_segmentation(seg_prev, d.shape[1:], order=1)
seg_reshaped = to_one_hot(seg_reshaped, classes)
d = np.vstack((d, seg_reshaped)).astype(np.float32)
"""There is a problem with python process communication that prevents us from communicating objects
larger than 2 GB between processes (basically when the length of the pickle string that will be sent is
communicated by the multiprocessing.Pipe object then the placeholder (I think) does not allow for long
enough strings (lol). This could be fixed by changing i to l (for long) but that would require manually
patching system python code. We circumvent that problem here by saving softmax_pred to a npy file that will
then be read (and finally deleted) by the Process. save_segmentation_nifti_from_softmax can take either
filename or np.ndarray and will handle this automatically"""
print(d.shape)
if np.prod(d.shape) > (2e9 / 4 * 0.85): # *0.85 just to be save, 4 because float32 is 4 bytes
print(
"This output is too large for python process-process communication. "
"Saving output temporarily to disk")
np.save(output_file[:-7] + ".npy", d)
d = output_file[:-7] + ".npy"
q.put((output_file, (d, dct)))
except KeyboardInterrupt:
raise KeyboardInterrupt
except Exception as e:
print("error in", l)
print(e)
q.put("end")
if len(errors_in) > 0:
print("There were some errors in the following cases:", errors_in)
print("These cases were ignored.")
else:
print("This worker has ended successfully, no errors to report")
# restore output
# sys.stdout = sys.__stdout__
def preprocess_multithreaded(trainer, list_of_lists, output_files, num_processes=2, segs_from_prev_stage=None):
if segs_from_prev_stage is None:
segs_from_prev_stage = [None] * len(list_of_lists)
num_processes = min(len(list_of_lists), num_processes)
classes = list(range(1, trainer.num_classes))
assert isinstance(trainer, nnUNetTrainer)
q = Queue(1)
processes = []
for i in range(num_processes):
pr = Process(target=preprocess_save_to_queue, args=(trainer.preprocess_patient, q,
list_of_lists[i::num_processes],
output_files[i::num_processes],
segs_from_prev_stage[i::num_processes],
classes, trainer.plans['transpose_forward']))
pr.start()
processes.append(pr)
try:
end_ctr = 0
while end_ctr != num_processes:
item = q.get()
if item == "end":
end_ctr += 1
continue
else:
yield item
finally:
for p in processes:
if p.is_alive():
p.terminate() # this should not happen but better safe than sorry right
p.join()
q.close()
def predict_cases(model, list_of_lists, output_filenames, folds, save_npz, num_threads_preprocessing,
num_threads_nifti_save, segs_from_prev_stage=None, do_tta=True, mixed_precision=True,
overwrite_existing=False,
all_in_gpu=False, step_size=0.5, checkpoint_name="model_final_checkpoint",
segmentation_export_kwargs: dict = None, disable_postprocessing: bool = False):
"""
:param segmentation_export_kwargs:
:param model: folder where the model is saved, must contain fold_x subfolders
:param list_of_lists: [[case0_0000.nii.gz, case0_0001.nii.gz], [case1_0000.nii.gz, case1_0001.nii.gz], ...]
:param output_filenames: [output_file_case0.nii.gz, output_file_case1.nii.gz, ...]
:param folds: default: (0, 1, 2, 3, 4) (but can also be 'all' or a subset of the five folds, for example use (0, )
for using only fold_0
:param save_npz: default: False
:param num_threads_preprocessing:
:param num_threads_nifti_save:
:param segs_from_prev_stage:
:param do_tta: default: True, can be set to False for a 8x speedup at the cost of a reduced segmentation quality
:param overwrite_existing: default: True
:param mixed_precision: if None then we take no action. If True/False we overwrite what the model has in its init
:return:
"""
assert len(list_of_lists) == len(output_filenames)
if segs_from_prev_stage is not None: assert len(segs_from_prev_stage) == len(output_filenames)
pool = Pool(num_threads_nifti_save)
results = []
cleaned_output_files = []
for o in output_filenames:
dr, f = os.path.split(o)
if len(dr) > 0:
maybe_mkdir_p(dr)
if not f.endswith(".nii.gz"):
f, _ = os.path.splitext(f)
f = f + ".nii.gz"
cleaned_output_files.append(join(dr, f))
if not overwrite_existing:
print("number of cases:", len(list_of_lists))
# if save_npz=True then we should also check for missing npz files
not_done_idx = [i for i, j in enumerate(cleaned_output_files) if (not isfile(j)) or (save_npz and not isfile(j[:-7] + '.npz'))]
cleaned_output_files = [cleaned_output_files[i] for i in not_done_idx]
list_of_lists = [list_of_lists[i] for i in not_done_idx]
if segs_from_prev_stage is not None:
segs_from_prev_stage = [segs_from_prev_stage[i] for i in not_done_idx]
print("number of cases that still need to be predicted:", len(cleaned_output_files))
print("emptying cuda cache")
torch.cuda.empty_cache()
print("loading parameters for folds,", folds)
trainer, params = load_model_and_checkpoint_files(model, folds, mixed_precision=mixed_precision,
checkpoint_name=checkpoint_name)
if segmentation_export_kwargs is None:
if 'segmentation_export_params' in trainer.plans.keys():
force_separate_z = trainer.plans['segmentation_export_params']['force_separate_z']
interpolation_order = trainer.plans['segmentation_export_params']['interpolation_order']
interpolation_order_z = trainer.plans['segmentation_export_params']['interpolation_order_z']
else:
force_separate_z = None
interpolation_order = 1
interpolation_order_z = 0
else:
force_separate_z = segmentation_export_kwargs['force_separate_z']
interpolation_order = segmentation_export_kwargs['interpolation_order']
interpolation_order_z = segmentation_export_kwargs['interpolation_order_z']
print("starting preprocessing generator")
preprocessing = preprocess_multithreaded(trainer, list_of_lists, cleaned_output_files, num_threads_preprocessing,
segs_from_prev_stage)
print("starting prediction...")
all_output_files = []
for preprocessed in preprocessing:
output_filename, (d, dct) = preprocessed
all_output_files.append(all_output_files)
if isinstance(d, str):
data = np.load(d)
os.remove(d)
d = data
print("predicting", output_filename)
trainer.load_checkpoint_ram(params[0], False)
softmax = trainer.predict_preprocessed_data_return_seg_and_softmax(
d, do_mirroring=do_tta, mirror_axes=trainer.data_aug_params['mirror_axes'], use_sliding_window=True,
step_size=step_size, use_gaussian=True, all_in_gpu=all_in_gpu,
mixed_precision=mixed_precision)[1]
for p in params[1:]:
trainer.load_checkpoint_ram(p, False)
softmax += trainer.predict_preprocessed_data_return_seg_and_softmax(
d, do_mirroring=do_tta, mirror_axes=trainer.data_aug_params['mirror_axes'], use_sliding_window=True,
step_size=step_size, use_gaussian=True, all_in_gpu=all_in_gpu,
mixed_precision=mixed_precision)[1]
if len(params) > 1:
softmax /= len(params)
transpose_forward = trainer.plans.get('transpose_forward')
if transpose_forward is not None:
transpose_backward = trainer.plans.get('transpose_backward')
softmax = softmax.transpose([0] + [i + 1 for i in transpose_backward])
if save_npz:
npz_file = output_filename[:-7] + ".npz"
else:
npz_file = None
if hasattr(trainer, 'regions_class_order'):
region_class_order = trainer.regions_class_order
else:
region_class_order = None
"""There is a problem with python process communication that prevents us from communicating objects
larger than 2 GB between processes (basically when the length of the pickle string that will be sent is
communicated by the multiprocessing.Pipe object then the placeholder (I think) does not allow for long
enough strings (lol). This could be fixed by changing i to l (for long) but that would require manually
patching system python code. We circumvent that problem here by saving softmax_pred to a npy file that will
then be read (and finally deleted) by the Process. save_segmentation_nifti_from_softmax can take either
filename or np.ndarray and will handle this automatically"""
bytes_per_voxel = 4
if all_in_gpu:
bytes_per_voxel = 2 # if all_in_gpu then the return value is half (float16)
if np.prod(softmax.shape) > (2e9 / bytes_per_voxel * 0.85): # * 0.85 just to be save
print(
"This output is too large for python process-process communication. Saving output temporarily to disk")
np.save(output_filename[:-7] + ".npy", softmax)
softmax = output_filename[:-7] + ".npy"
# save_segmentation_nifti_from_softmax(softmax, output_filename, dct, interpolation_order, region_class_order,
# None, None,
# npz_file, None, force_separate_z, interpolation_order_z)
results.append(pool.starmap_async(save_segmentation_nifti_from_softmax,
((softmax, output_filename, dct, interpolation_order, region_class_order,
None, None,
npz_file, None, force_separate_z, interpolation_order_z),)
))
print("inference done. Now waiting for the segmentation export to finish...")
_ = [i.get() for i in results]
# now apply postprocessing
# first load the postprocessing properties if they are present. Else raise a well visible warning
if not disable_postprocessing:
results = []
pp_file = join(model, "postprocessing.json")
if isfile(pp_file):
print("postprocessing...")
shutil.copy(pp_file, os.path.abspath(os.path.dirname(output_filenames[0])))
# for_which_classes stores for which of the classes everything but the largest connected component needs to be
# removed
for_which_classes, min_valid_obj_size = load_postprocessing(pp_file)
results.append(pool.starmap_async(load_remove_save,
zip(output_filenames, output_filenames,
[for_which_classes] * len(output_filenames),
[min_valid_obj_size] * len(output_filenames))))
_ = [i.get() for i in results]
else:
print("WARNING! Cannot run postprocessing because the postprocessing file is missing. Make sure to run "
"consolidate_folds in the output folder of the model first!\nThe folder you need to run this in is "
"%s" % model)
pool.close()
pool.join()
def predict_cases_fast(model, list_of_lists, output_filenames, folds, num_threads_preprocessing,
num_threads_nifti_save, segs_from_prev_stage=None, do_tta=True, mixed_precision=True,
overwrite_existing=False,
all_in_gpu=False, step_size=0.5, checkpoint_name="model_final_checkpoint",
segmentation_export_kwargs: dict = None, disable_postprocessing: bool = False):
assert len(list_of_lists) == len(output_filenames)
if segs_from_prev_stage is not None: assert len(segs_from_prev_stage) == len(output_filenames)
pool = Pool(num_threads_nifti_save)
results = []
cleaned_output_files = []
for o in output_filenames:
dr, f = os.path.split(o)
if len(dr) > 0:
maybe_mkdir_p(dr)
if not f.endswith(".nii.gz"):
f, _ = os.path.splitext(f)
f = f + ".nii.gz"
cleaned_output_files.append(join(dr, f))
if not overwrite_existing:
print("number of cases:", len(list_of_lists))
not_done_idx = [i for i, j in enumerate(cleaned_output_files) if not isfile(j)]
cleaned_output_files = [cleaned_output_files[i] for i in not_done_idx]
list_of_lists = [list_of_lists[i] for i in not_done_idx]
if segs_from_prev_stage is not None:
segs_from_prev_stage = [segs_from_prev_stage[i] for i in not_done_idx]
print("number of cases that still need to be predicted:", len(cleaned_output_files))
print("emptying cuda cache")
torch.cuda.empty_cache()
print("loading parameters for folds,", folds)
trainer, params = load_model_and_checkpoint_files(model, folds, mixed_precision=mixed_precision,
checkpoint_name=checkpoint_name)
if segmentation_export_kwargs is None:
if 'segmentation_export_params' in trainer.plans.keys():
force_separate_z = trainer.plans['segmentation_export_params']['force_separate_z']
interpolation_order = trainer.plans['segmentation_export_params']['interpolation_order']
interpolation_order_z = trainer.plans['segmentation_export_params']['interpolation_order_z']
else:
force_separate_z = None
interpolation_order = 1
interpolation_order_z = 0
else:
force_separate_z = segmentation_export_kwargs['force_separate_z']
interpolation_order = segmentation_export_kwargs['interpolation_order']
interpolation_order_z = segmentation_export_kwargs['interpolation_order_z']
print("starting preprocessing generator")
preprocessing = preprocess_multithreaded(trainer, list_of_lists, cleaned_output_files, num_threads_preprocessing,
segs_from_prev_stage)
print("starting prediction...")
for preprocessed in preprocessing:
print("getting data from preprocessor")
output_filename, (d, dct) = preprocessed
print("got something")
if isinstance(d, str):
print("what I got is a string, so I need to load a file")
data = np.load(d)
os.remove(d)
d = data
# preallocate the output arrays
# same dtype as the return value in predict_preprocessed_data_return_seg_and_softmax (saves time)
softmax_aggr = None # np.zeros((trainer.num_classes, *d.shape[1:]), dtype=np.float16)
all_seg_outputs = np.zeros((len(params), *d.shape[1:]), dtype=int)
print("predicting", output_filename)
for i, p in enumerate(params):
trainer.load_checkpoint_ram(p, False)
res = trainer.predict_preprocessed_data_return_seg_and_softmax(d, do_mirroring=do_tta,
mirror_axes=trainer.data_aug_params['mirror_axes'],
use_sliding_window=True,
step_size=step_size, use_gaussian=True,
all_in_gpu=all_in_gpu,
mixed_precision=mixed_precision)
if len(params) > 1:
# otherwise we dont need this and we can save ourselves the time it takes to copy that
print("aggregating softmax")
if softmax_aggr is None:
softmax_aggr = res[1]
else:
softmax_aggr += res[1]
all_seg_outputs[i] = res[0]
print("obtaining segmentation map")
if len(params) > 1:
# we dont need to normalize the softmax by 1 / len(params) because this would not change the outcome of the argmax
seg = softmax_aggr.argmax(0)
else:
seg = all_seg_outputs[0]
print("applying transpose_backward")
transpose_forward = trainer.plans.get('transpose_forward')
if transpose_forward is not None:
transpose_backward = trainer.plans.get('transpose_backward')
seg = seg.transpose([i for i in transpose_backward])
if hasattr(trainer, 'regions_class_order'):
region_class_order = trainer.regions_class_order
else:
region_class_order = None
assert region_class_order is None, "predict_cases_fast can only work with regular softmax predictions " \
"and is therefore unable to handle trainer classes with region_class_order"
print("initializing segmentation export")
results.append(pool.starmap_async(save_segmentation_nifti,
((seg, output_filename, dct, interpolation_order, force_separate_z,
interpolation_order_z),)
))
print("done")
print("inference done. Now waiting for the segmentation export to finish...")
_ = [i.get() for i in results]
# now apply postprocessing
# first load the postprocessing properties if they are present. Else raise a well visible warning
if not disable_postprocessing:
results = []
pp_file = join(model, "postprocessing.json")
if isfile(pp_file):
print("postprocessing...")
shutil.copy(pp_file, os.path.dirname(output_filenames[0]))
# for_which_classes stores for which of the classes everything but the largest connected component needs to be
# removed
for_which_classes, min_valid_obj_size = load_postprocessing(pp_file)
results.append(pool.starmap_async(load_remove_save,
zip(output_filenames, output_filenames,
[for_which_classes] * len(output_filenames),
[min_valid_obj_size] * len(output_filenames))))
_ = [i.get() for i in results]
else:
print("WARNING! Cannot run postprocessing because the postprocessing file is missing. Make sure to run "
"consolidate_folds in the output folder of the model first!\nThe folder you need to run this in is "
"%s" % model)
pool.close()
pool.join()
def predict_cases_fastest(model, list_of_lists, output_filenames, folds, num_threads_preprocessing,
num_threads_nifti_save, segs_from_prev_stage=None, do_tta=True, mixed_precision=True,
overwrite_existing=False, all_in_gpu=False, step_size=0.5,
checkpoint_name="model_final_checkpoint", disable_postprocessing: bool = False):
assert len(list_of_lists) == len(output_filenames)
if segs_from_prev_stage is not None: assert len(segs_from_prev_stage) == len(output_filenames)
pool = Pool(num_threads_nifti_save)
results = []
cleaned_output_files = []
for o in output_filenames:
dr, f = os.path.split(o)
if len(dr) > 0:
maybe_mkdir_p(dr)
if not f.endswith(".nii.gz"):
f, _ = os.path.splitext(f)
f = f + ".nii.gz"
cleaned_output_files.append(join(dr, f))
if not overwrite_existing:
print("number of cases:", len(list_of_lists))
not_done_idx = [i for i, j in enumerate(cleaned_output_files) if not isfile(j)]
cleaned_output_files = [cleaned_output_files[i] for i in not_done_idx]
list_of_lists = [list_of_lists[i] for i in not_done_idx]
if segs_from_prev_stage is not None:
segs_from_prev_stage = [segs_from_prev_stage[i] for i in not_done_idx]
print("number of cases that still need to be predicted:", len(cleaned_output_files))
print("emptying cuda cache")
torch.cuda.empty_cache()
print("loading parameters for folds,", folds)
trainer, params = load_model_and_checkpoint_files(model, folds, mixed_precision=mixed_precision,
checkpoint_name=checkpoint_name)
print("starting preprocessing generator")
preprocessing = preprocess_multithreaded(trainer, list_of_lists, cleaned_output_files, num_threads_preprocessing,
segs_from_prev_stage)
print("starting prediction...")
for preprocessed in preprocessing:
print("getting data from preprocessor")
output_filename, (d, dct) = preprocessed
print("got something")
if isinstance(d, str):
print("what I got is a string, so I need to load a file")
data = np.load(d)
os.remove(d)
d = data
# preallocate the output arrays
# same dtype as the return value in predict_preprocessed_data_return_seg_and_softmax (saves time)
all_softmax_outputs = np.zeros((len(params), trainer.num_classes, *d.shape[1:]), dtype=np.float16)
all_seg_outputs = np.zeros((len(params), *d.shape[1:]), dtype=int)
print("predicting", output_filename)
for i, p in enumerate(params):
trainer.load_checkpoint_ram(p, False)
res = trainer.predict_preprocessed_data_return_seg_and_softmax(d, do_mirroring=do_tta,
mirror_axes=trainer.data_aug_params['mirror_axes'],
use_sliding_window=True,
step_size=step_size, use_gaussian=True,
all_in_gpu=all_in_gpu,
mixed_precision=mixed_precision)
if len(params) > 1:
# otherwise we dont need this and we can save ourselves the time it takes to copy that
all_softmax_outputs[i] = res[1]
all_seg_outputs[i] = res[0]
if hasattr(trainer, 'regions_class_order'):
region_class_order = trainer.regions_class_order
else:
region_class_order = None
assert region_class_order is None, "predict_cases_fastest can only work with regular softmax predictions " \
"and is therefore unable to handle trainer classes with region_class_order"
print("aggregating predictions")
if len(params) > 1:
softmax_mean = np.mean(all_softmax_outputs, 0)
seg = softmax_mean.argmax(0)
else:
seg = all_seg_outputs[0]
print("applying transpose_backward")
transpose_forward = trainer.plans.get('transpose_forward')
if transpose_forward is not None:
transpose_backward = trainer.plans.get('transpose_backward')
seg = seg.transpose([i for i in transpose_backward])
print("initializing segmentation export")
results.append(pool.starmap_async(save_segmentation_nifti,
((seg, output_filename, dct, 0, None),)
))
print("done")
print("inference done. Now waiting for the segmentation export to finish...")
_ = [i.get() for i in results]
# now apply postprocessing
# first load the postprocessing properties if they are present. Else raise a well visible warning
if not disable_postprocessing:
results = []
pp_file = join(model, "postprocessing.json")
if isfile(pp_file):
print("postprocessing...")
shutil.copy(pp_file, os.path.dirname(output_filenames[0]))
# for_which_classes stores for which of the classes everything but the largest connected component needs to be
# removed
for_which_classes, min_valid_obj_size = load_postprocessing(pp_file)
results.append(pool.starmap_async(load_remove_save,
zip(output_filenames, output_filenames,
[for_which_classes] * len(output_filenames),
[min_valid_obj_size] * len(output_filenames))))
_ = [i.get() for i in results]
else:
print("WARNING! Cannot run postprocessing because the postprocessing file is missing. Make sure to run "
"consolidate_folds in the output folder of the model first!\nThe folder you need to run this in is "
"%s" % model)
pool.close()
pool.join()
def check_input_folder_and_return_caseIDs(input_folder, expected_num_modalities):
print("This model expects %d input modalities for each image" % expected_num_modalities)
files = subfiles(input_folder, suffix=".nii.gz", join=False, sort=True)
maybe_case_ids = np.unique([i[:-12] for i in files])
remaining = deepcopy(files)
missing = []
assert len(files) > 0, "input folder did not contain any images (expected to find .nii.gz file endings)"
# now check if all required files are present and that no unexpected files are remaining
for c in maybe_case_ids:
for n in range(expected_num_modalities):
expected_output_file = c + "_%04.0d.nii.gz" % n
if not isfile(join(input_folder, expected_output_file)):
missing.append(expected_output_file)
else:
remaining.remove(expected_output_file)
print("Found %d unique case ids, here are some examples:" % len(maybe_case_ids),
np.random.choice(maybe_case_ids, min(len(maybe_case_ids), 10)))
print("If they don't look right, make sure to double check your filenames. They must end with _0000.nii.gz etc")
if len(remaining) > 0:
print("found %d unexpected remaining files in the folder. Here are some examples:" % len(remaining),
np.random.choice(remaining, min(len(remaining), 10)))
if len(missing) > 0:
print("Some files are missing:")
print(missing)
raise RuntimeError("missing files in input_folder")
return maybe_case_ids
def predict_from_folder(model: str, input_folder: str, output_folder: str, folds: Union[Tuple[int], List[int]],
save_npz: bool, num_threads_preprocessing: int, num_threads_nifti_save: int,
lowres_segmentations: Union[str, None],
part_id: int, num_parts: int, tta: bool, mixed_precision: bool = True,
overwrite_existing: bool = True, mode: str = 'normal', overwrite_all_in_gpu: bool = None,
step_size: float = 0.5, checkpoint_name: str = "model_final_checkpoint",
segmentation_export_kwargs: dict = None, disable_postprocessing: bool = False):
"""
here we use the standard naming scheme to generate list_of_lists and output_files needed by predict_cases
:param model:
:param input_folder:
:param output_folder:
:param folds:
:param save_npz:
:param num_threads_preprocessing:
:param num_threads_nifti_save:
:param lowres_segmentations:
:param part_id:
:param num_parts:
:param tta:
:param mixed_precision:
:param overwrite_existing: if not None then it will be overwritten with whatever is in there. None is default (no overwrite)
:return:
"""
maybe_mkdir_p(output_folder)
shutil.copy(join(model, 'plans.pkl'), output_folder)
assert isfile(join(model, "plans.pkl")), "Folder with saved model weights must contain a plans.pkl file"
expected_num_modalities = load_pickle(join(model, "plans.pkl"))['num_modalities']
# check input folder integrity
case_ids = check_input_folder_and_return_caseIDs(input_folder, expected_num_modalities)
output_files = [join(output_folder, i + ".nii.gz") for i in case_ids]
all_files = subfiles(input_folder, suffix=".nii.gz", join=False, sort=True)
list_of_lists = [[join(input_folder, i) for i in all_files if i[:len(j)].startswith(j) and
len(i) == (len(j) + 12)] for j in case_ids]
if lowres_segmentations is not None:
assert isdir(lowres_segmentations), "if lowres_segmentations is not None then it must point to a directory"
lowres_segmentations = [join(lowres_segmentations, i + ".nii.gz") for i in case_ids]
assert all([isfile(i) for i in lowres_segmentations]), "not all lowres_segmentations files are present. " \
"(I was searching for case_id.nii.gz in that folder)"
lowres_segmentations = lowres_segmentations[part_id::num_parts]
else:
lowres_segmentations = None
if mode == "normal":
if overwrite_all_in_gpu is None:
all_in_gpu = False
else:
all_in_gpu = overwrite_all_in_gpu
return predict_cases(model, list_of_lists[part_id::num_parts], output_files[part_id::num_parts], folds,
save_npz, num_threads_preprocessing, num_threads_nifti_save, lowres_segmentations, tta,
mixed_precision=mixed_precision, overwrite_existing=overwrite_existing,
all_in_gpu=all_in_gpu,
step_size=step_size, checkpoint_name=checkpoint_name,
segmentation_export_kwargs=segmentation_export_kwargs,
disable_postprocessing=disable_postprocessing)
elif mode == "fast":
if overwrite_all_in_gpu is None:
all_in_gpu = False
else:
all_in_gpu = overwrite_all_in_gpu
assert save_npz is False
return predict_cases_fast(model, list_of_lists[part_id::num_parts], output_files[part_id::num_parts], folds,
num_threads_preprocessing, num_threads_nifti_save, lowres_segmentations,
tta, mixed_precision=mixed_precision, overwrite_existing=overwrite_existing,
all_in_gpu=all_in_gpu,
step_size=step_size, checkpoint_name=checkpoint_name,
segmentation_export_kwargs=segmentation_export_kwargs,
disable_postprocessing=disable_postprocessing)
elif mode == "fastest":
if overwrite_all_in_gpu is None:
all_in_gpu = False
else:
all_in_gpu = overwrite_all_in_gpu
assert save_npz is False
return predict_cases_fastest(model, list_of_lists[part_id::num_parts], output_files[part_id::num_parts], folds,
num_threads_preprocessing, num_threads_nifti_save, lowres_segmentations,
tta, mixed_precision=mixed_precision, overwrite_existing=overwrite_existing,
all_in_gpu=all_in_gpu,
step_size=step_size, checkpoint_name=checkpoint_name,
disable_postprocessing=disable_postprocessing)
else:
raise ValueError("unrecognized mode. Must be normal, fast or fastest")
if __name__ == "__main__":
parser = argparse.ArgumentParser()
parser.add_argument("-i", '--input_folder', help="Must contain all modalities for each patient in the correct"
" order (same as training). Files must be named "
"CASENAME_XXXX.nii.gz where XXXX is the modality "
"identifier (0000, 0001, etc)", required=True)
parser.add_argument('-o', "--output_folder", required=True, help="folder for saving predictions")
parser.add_argument('-m', '--model_output_folder',
help='model output folder. Will automatically discover the folds '
'that were '
'run and use those as an ensemble', required=True)
parser.add_argument('-f', '--folds', nargs='+', default='None', help="folds to use for prediction. Default is None "
"which means that folds will be detected "
"automatically in the model output folder")
parser.add_argument('-z', '--save_npz', required=False, action='store_true', help="use this if you want to ensemble"
" these predictions with those of"
" other models. Softmax "
"probabilities will be saved as "
"compresed numpy arrays in "
"output_folder and can be merged "
"between output_folders with "
"merge_predictions.py")
parser.add_argument('-l', '--lowres_segmentations', required=False, default='None', help="if model is the highres "
"stage of the cascade then you need to use -l to specify where the segmentations of the "
"corresponding lowres unet are. Here they are required to do a prediction")
parser.add_argument("--part_id", type=int, required=False, default=0, help="Used to parallelize the prediction of "
"the folder over several GPUs. If you "
"want to use n GPUs to predict this "
"folder you need to run this command "
"n times with --part_id=0, ... n-1 and "
"--num_parts=n (each with a different "
"GPU (for example via "
"CUDA_VISIBLE_DEVICES=X)")
parser.add_argument("--num_parts", type=int, required=False, default=1,
help="Used to parallelize the prediction of "
"the folder over several GPUs. If you "
"want to use n GPUs to predict this "
"folder you need to run this command "
"n times with --part_id=0, ... n-1 and "
"--num_parts=n (each with a different "
"GPU (via "
"CUDA_VISIBLE_DEVICES=X)")
parser.add_argument("--num_threads_preprocessing", required=False, default=6, type=int, help=
"Determines many background processes will be used for data preprocessing. Reduce this if you "
"run into out of memory (RAM) problems. Default: 6")
parser.add_argument("--num_threads_nifti_save", required=False, default=2, type=int, help=
"Determines many background processes will be used for segmentation export. Reduce this if you "
"run into out of memory (RAM) problems. Default: 2")
parser.add_argument("--tta", required=False, type=int, default=1, help="Set to 0 to disable test time data "
"augmentation (speedup of factor "
"4(2D)/8(3D)), "
"lower quality segmentations")
parser.add_argument("--overwrite_existing", required=False, type=int, default=1, help="Set this to 0 if you need "
"to resume a previous "
"prediction. Default: 1 "
"(=existing segmentations "
"in output_folder will be "
"overwritten)")
parser.add_argument("--mode", type=str, default="normal", required=False)
parser.add_argument("--all_in_gpu", type=str, default="None", required=False, help="can be None, False or True")
parser.add_argument("--step_size", type=float, default=0.5, required=False, help="don't touch")
# parser.add_argument("--interp_order", required=False, default=3, type=int,
# help="order of interpolation for segmentations, has no effect if mode=fastest")
# parser.add_argument("--interp_order_z", required=False, default=0, type=int,
# help="order of interpolation along z is z is done differently")
# parser.add_argument("--force_separate_z", required=False, default="None", type=str,
# help="force_separate_z resampling. Can be None, True or False, has no effect if mode=fastest")
parser.add_argument('--disable_mixed_precision', default=False, action='store_true', required=False,
help='Predictions are done with mixed precision by default. This improves speed and reduces '
'the required vram. If you want to disable mixed precision you can set this flag. Note '
'that this is not recommended (mixed precision is ~2x faster!)')
args = parser.parse_args()
input_folder = args.input_folder
output_folder = args.output_folder
part_id = args.part_id
num_parts = args.num_parts
model = args.model_output_folder
folds = args.folds
save_npz = args.save_npz
lowres_segmentations = args.lowres_segmentations
num_threads_preprocessing = args.num_threads_preprocessing
num_threads_nifti_save = args.num_threads_nifti_save
tta = args.tta
step_size = args.step_size
# interp_order = args.interp_order
# interp_order_z = args.interp_order_z
# force_separate_z = args.force_separate_z
# if force_separate_z == "None":
# force_separate_z = None
# elif force_separate_z == "False":
# force_separate_z = False
# elif force_separate_z == "True":
# force_separate_z = True
# else:
# raise ValueError("force_separate_z must be None, True or False. Given: %s" % force_separate_z)
overwrite = args.overwrite_existing
mode = args.mode
all_in_gpu = args.all_in_gpu
if lowres_segmentations == "None":
lowres_segmentations = None
if isinstance(folds, list):
if folds[0] == 'all' and len(folds) == 1:
pass
else:
folds = [int(i) for i in folds]
elif folds == "None":
folds = None
else:
raise ValueError("Unexpected value for argument folds")
if tta == 0:
tta = False
elif tta == 1:
tta = True
else:
raise ValueError("Unexpected value for tta, Use 1 or 0")
if overwrite == 0:
overwrite = False
elif overwrite == 1:
overwrite = True
else:
raise ValueError("Unexpected value for overwrite, Use 1 or 0")
assert all_in_gpu in ['None', 'False', 'True']
if all_in_gpu == "None":
all_in_gpu = None
elif all_in_gpu == "True":
all_in_gpu = True
elif all_in_gpu == "False":
all_in_gpu = False
predict_from_folder(model, input_folder, output_folder, folds, save_npz, num_threads_preprocessing,
num_threads_nifti_save, lowres_segmentations, part_id, num_parts, tta,
mixed_precision=not args.disable_mixed_precision,
overwrite_existing=overwrite, mode=mode, overwrite_all_in_gpu=all_in_gpu, step_size=step_size)
nnunet的测试分为两个阶段:
| 阶段 | 名称 | 作用 | 是否自动完成 |
|---|---|---|---|
| 推理(Inference / Prediction) | nnUNet_predict |
将模型应用于测试图像,生成预测的 .nii.gz 分割结果 |
需手动运行 |
| 评估(Evaluation) | nnUNet_evaluate_folder 或自定义脚本 |
将预测结果与 ground truth 比较,计算 Dice、HD95 等指标 | 需手动运行 |
推理命令:
# 1. 设置 nnU-Net 所需的环境变量
export nnUNet_raw_data_base="/xujiheng/Synapse/nnUNet/nnUNet/nnUNetFrame/DATASET/nnUNet_raw"
export nnUNet_preprocessed="/xujiheng/Synapse/nnUNet/nnUNet/nnUNetFrame/DATASET/nnUNet_preprocessed"
export RESULTS_FOLDER="/xujiheng/Synapse/nnUNet/nnUNet/nnUNetFrame/DATASET/nnUNet_trained_models"
# 2. 设置预测结果保存目录
PRED_DIR="/xujiheng/Synapse/nnUNet/nnUNet/nnUNetFrame/DATASET/nnUNet_trained_models/test_predictions_Task500"
mkdir -p $PRED_DIR
# 3. 运行推理(使用 all fold + model_best)
nnUNet_predict \
-i $nnUNet_raw_data_base/nnUNet_raw_data/Task500_Synapse/imagesTs \
-o $PRED_DIR \
-t 500 \
-m 3d_fullres \
-tr nnUNetTrainerV2 \
-f all \
-chk model_best解释:
(1)nnUNet_raw_data_base:原始数据的根目录,也就是你的 imagesTr/, labelsTr/, imagesTs/ 所在位置。
nnUNet_preprocessed:nnU-Net 在训练前生成的预处理数据保存路径(比如 resampled、normalized 数据)。
RESULTS_FOLDER:训练模型和预测结果的默认保存目录。
(2)PRED_DIR:把预测的结果保存到哪里。
mkdir -p:确保目录存在,如果不存在就创建。所有推理生成的 .nii.gz 文件都会保存到这里。
(3)参数
| 参数 | 说明 |
|---|---|
-i |
输入目录,指定为测试集图像目录 imagesTs |
-o |
输出目录,使用之前定义的环境变量 $PRED_DIR |
-t |
Task ID,设为 500,对应 Synapse 数据集(Task500) |
-m |
模型类型,使用 3d_fullres(完整分辨率 3D 模型) |
-tr |
Trainer 类,指定为 nnUNetTrainerV2(常用训练器) |
-f |
Fold,设为 all,表示使用所有训练好的 fold 模型进行预测并集成(ensemble) |
-chk |
检查点,使用 model_best(最优模型权重,而非最后训练轮次的权重) |
推理后的结果目录内容如下:
评估命令:
nnUNet_evaluate_folder -ref /xujiheng/Synapse/nnUNet/nnUNet/nnUNetFrame/DATASET/nnUNet_raw/nnUNet_raw_data/Task500_Synapse/labelsTs -pred /xujiheng/Synapse/nnUNet/nnUNet/nnUNetFrame/DATASET/nnUNet_trained_models/test_predictions_Task500 -l 1 2 3 4 5 6 7 8
解释:
| 参数 | 说明 |
|---|---|
-ref |
测试集真实标签路径 |
-pred |
nnU-Net 预测结果路径(即上一步生成的 $PRED_DIR) |
-l |
指定需要评估的标签 ID,此处为 1-8,对应 8 个器官(不包含背景类别 0) |
补充
现在来到正题,之前提到过nnunet适合作为一个框架去使用,所以接下来我要做的是,使用它去跑其他的网络结构。
即在不修改 nnU-Net 核心架构的前提下,通过模块化扩展方式,使用 nnU-Net 的完整训练流水线(数据预处理 → 训练 → 推理),但将默认的 Generic_UNet 替换为自定义网络(如 SwinUNet)。
下面是要干的事情:
| 操作 | 内容 | 是否必须 |
|---|---|---|
| 添加 | 自定义网络文件(如 swin_unet.py) |
是 |
| 添加 | 新 Trainer 文件:nnUNetTrainerV2_SwinUNet.py |
是 |
| 修改 | 在新 Trainer 中重写 initialize_network() |
是 |
| 保持 | 不动 run_training.py |
是(正确做法) |
| 可选 | 重写 initialize() 调整 batch size / patch size |
否 |
| 可选 | 重写 initialize_loss() 换损失函数 |
否 |
| 运行 | 命令行指定你的 Trainer 名字 | 是 |
添加网络结构.py文件,对应也添加一个Trainer.py,这样更加规范有条理。
关于这个扩展内容,博主会尝试,后续会继续更新。
更多推荐
 已为社区贡献3条内容
已为社区贡献3条内容








所有评论(0)