Halcon学习---案例实战(5)--药片检测(高斯混合模型分类)
check_blister_mixed.hdev说明:这个案例很有意思,既运用了Blob分析定位+决策判断,也运用了高斯混合模型进行训练分类;把这个案例反复读几遍,抠细节,体会整个解题思路;* This example demonstrates an application from the pharmaceutical* industry. The task is to check the co
·
check_blister_mixed.hdev
说明:
这个案例很有意思,既运用了Blob分析定位+决策判断,也运用了高斯混合模型进行训练分类;把这个案例反复读几遍,抠细节,体会整个解题思路;
* This example demonstrates an application from the pharmaceutical
* industry. The task is to check the content of manually filled
* blisters. The first image (reference) shows the combination, that
* is requested. The subsequent images are then checked for this
* setup drawn from the reference image. To perform this task we use
* the gmm classifier, which will be trained with the reference image
* in the beginning and is then used to classify the different pill
* types in subsequent blister images.
*
dev_close_window ()
read_image (Image, 'blister/blister_mixed_reference')
dev_open_window_fit_image (Image, 0, 0, -1, -1, WindowHandle)
set_display_font (WindowHandle, 14, 'mono', 'true', 'false')
dev_update_off ()
dev_display (Image)
dev_set_draw ('margin')
*
* First, we extract the content of the blister and pass
* this information on to the gmm classifier
disp_message (WindowHandle, 'Train gmm classifier on pill types', 'window', -1, -1, 'black', 'true')
*这是一个特征提取的函数,halcon自带的,里面的思路比较简单,分别提取几个药片的主要特征,提供给gmm
*学习,三种主要特征,三个类,以及后面用于仿射变换的参数
extract_pill_types (Image, Chambers, ChambersUnion, Classes, PhiRef, RowRef, ColumnRef, PillTypeCount)
NumClasses := |PillTypeCount|
*创建一个高斯混合模型
create_class_gmm (3, 3, [1,5], 'spherical', 'normalization', 10, 42, GMMHandle)
*把图片和对应的特征放到模型中
add_samples_image_class_gmm (Image, Classes, GMMHandle, 0)
*训练这个模型
train_class_gmm (GMMHandle, 100, 0.001, 'training', 0.0001, Centers, Iter)
*
*
* Then, the subsequent blisters are tested for their right combination
Count := 12
for FileIndex := 1 to Count by 1
*
* Align image read
read_image (Image, 'blister/blister_mixed_' + FileIndex$'02')
threshold (Image, Region, 90, 255)
connection (Region, ConnectedRegions)
select_shape (ConnectedRegions, SelectedRegions, 'area', 'and', 5000, 9999999)
shape_trans (SelectedRegions, RegionTrans, 'convex')
orientation_region (RegionTrans, Phi)
if (abs(Phi) > rad(90))
Phi := rad(180) + Phi
endif
area_center (RegionTrans, Area1, Row, Column)
vector_angle_to_rigid (Row, Column, Phi, RowRef, ColumnRef, PhiRef, HomMat2D)
affine_trans_image (Image, ImageAffinTrans, HomMat2D, 'constant', 'false')
*上面都是在对一张新图进行Blob分析+定位
*下面这句表示要抠出分类的区域
reduce_domain (ImageAffinTrans, ChambersUnion, ImageReduced)
decompose3 (ImageAffinTrans, ImageR, ImageG, ImageB)
*
* Classify pill type for each chamber
*用训练好的模型对区域进行分类
classify_image_class_gmm (ImageReduced, ClassRegions, GMMHandle, 0.005)
*
count_obj (ClassRegions, Number)
gen_empty_obj (FinalClasses)
connection (Chambers, ChambersRemaining)
for Index := Number to 1 by -1
dev_clear_window ()
*从分类的结果中选出一个指定类
select_obj (ClassRegions, Region, Index)
*与抠出来的总区域做交集--确保得到的是药品区域
intersection (ChambersRemaining, Region, Region)
*根据面积和宽度筛选掉一部分--这时候太长和太短以及面积太小和太大的均筛选掉了
select_shape (Region, PillsOfOneType, ['area','width'], 'and', [200,40], [3000,68])
*用整个大区域减去特征区域
difference (ChambersUnion, PillsOfOneType, RegionDifference)
*打散
connection (RegionDifference, ConnectedRegions)
*根据面积选出已经推断的区域
select_shape (ConnectedRegions, SelectedRegions, 'area', 'and', 0, 7868)
*凸性转换(这里相当于填充)
shape_trans (SelectedRegions, SelectedRegions, 'convex')
*某个类别的所有药品联合起来
union1 (SelectedRegions, SelectedRegions)
*差分得到还没有推断的区域
difference (ChambersRemaining, SelectedRegions, ChambersRemaining)
*推断的结果开始叠加起来
concat_obj (SelectedRegions, FinalClasses, FinalClasses)
endfor
*
*
* Check for right combination
gen_empty_obj (MissingPills)
gen_empty_obj (WrongPills)
gen_empty_obj (WrongNumberOfPills)
*差分要推断的区域和最终的结果区域,如果有多的说明是错误的或缺少的
difference (ChambersUnion, FinalClasses, LeftOvers)
area_center (LeftOvers, Area, Row1, Column1)
if (Area > 0)
connection (LeftOvers, LeftOvers)
count_obj (LeftOvers, Number)
for Index := 1 to Number by 1
select_obj (LeftOvers, ObjectSelected, Index)
*计算图片中某个区域的均值和方差
intensity (ObjectSelected, ImageB, Mean, Deviation)
if (Deviation > 40)
concat_obj (WrongPills, ObjectSelected, WrongPills)
else
concat_obj (MissingPills, ObjectSelected, MissingPills)
endif
endfor
endif
*
* Compute histogram
CountFinalClass := []
for Index := 1 to NumClasses by 1
select_obj (FinalClasses, ObjectSelected, Index)
connection (ObjectSelected, ObjectSelected)
count_obj (ObjectSelected, Size)
CountFinalClass := [CountFinalClass,Size]
endfor
*
* Display classification results and output allover statistic
display_results (ImageAffinTrans, LeftOvers, FinalClasses, WrongPills, CountFinalClass, PillTypeCount, WindowHandle)
if (FileIndex < Count)
disp_continue_message (WindowHandle, 'black', 'true')
endif
stop ()
endfor
*
*
* Clear classifier handle
clear_class_gmm (GMMHandle)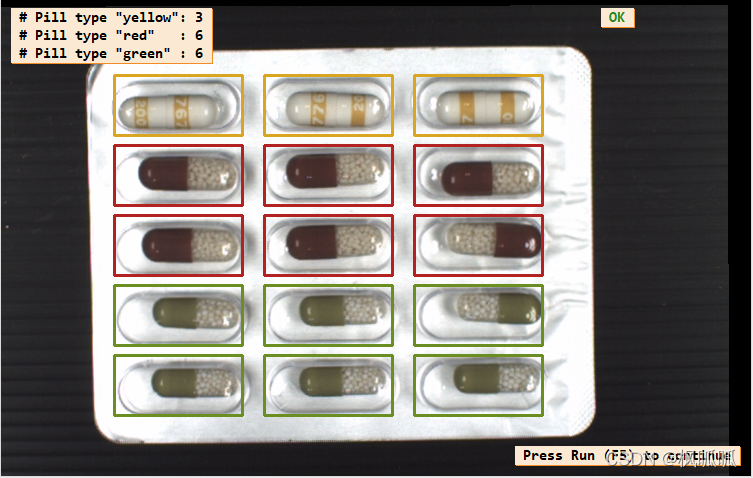



更多推荐
 已为社区贡献10条内容
已为社区贡献10条内容








所有评论(0)